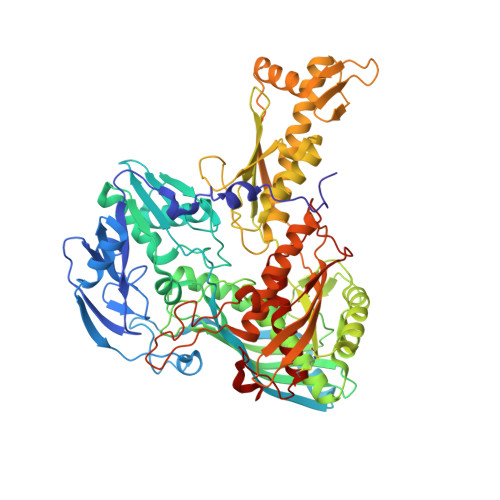
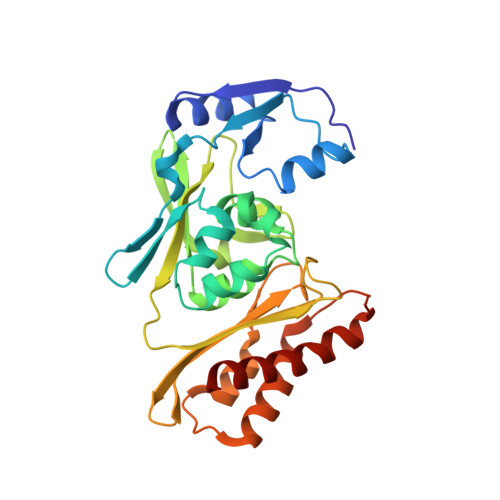
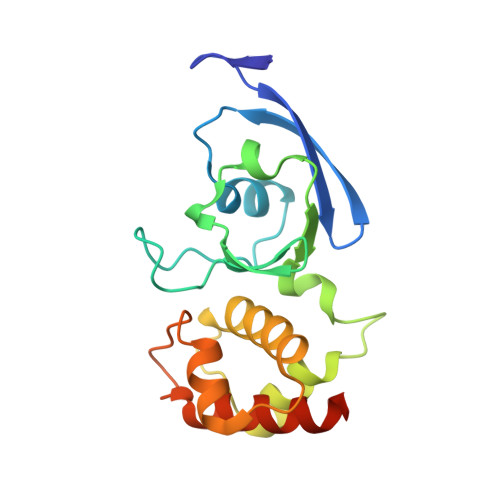
Archaeal Mo-Containing Glyceraldehyde Oxidoreductase Isozymes Exhibit Diverse Substrate Specificities through Unique Subunit Assemblies.
Wakagi, T., Nishimasu, H., Miyake, M., Fushinobu, S.(2016) PLoS One 11: e0147333-e0147333
- PubMed: 26808202
- DOI: https://doi.org/10.1371/journal.pone.0147333
- Primary Citation of Related Structures:
4ZOH - PubMed Abstract:
Archaea use glycolytic pathways distinct from those found in bacteria and eukaryotes, where unique enzymes catalyze each reaction step. In this study, we isolated three isozymes of glyceraldehyde oxidoreductase (GAOR1, GAOR2 and GAOR3) from the thermoacidophilic archaeon Sulfolobus tokodaii. GAOR1-3 belong to the xanthine oxidoreductase superfamily, and are composed of a molybdo-pyranopterin subunit (L), a flavin subunit (M), and an iron-sulfur subunit (S), forming an LMS hetero-trimer unit. We found that GAOR1 is a tetramer of the STK17810/STK17830/STK17820 hetero-trimer, GAOR2 is a dimer of the STK23390/STK05620/STK05610 hetero-trimer, and GAOR3 is the STK24840/STK05620/STK05610 hetero-trimer. GAOR1-3 exhibited diverse substrate specificities for their electron donors and acceptors, due to their different L-subunits, and probably participate in the non-phosphorylative Entner-Doudoroff glycolytic pathway. We determined the crystal structure of GAOR2, as the first three-dimensional structure of an archaeal molybdenum-containing hydroxylase, to obtain structural insights into their substrate specificities and subunit assemblies. The gene arrangement and the crystal structure suggested that the M/S-complex serves as a structural scaffold for the binding of the L-subunit, to construct the three enzymes with different specificities. Collectively, our findings illustrate a novel principle of a prokaryotic multicomponent isozyme system.
Organizational Affiliation:
Department of Biotechnology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Yayoi, Bunkyo-ku, Tokyo, Japan.