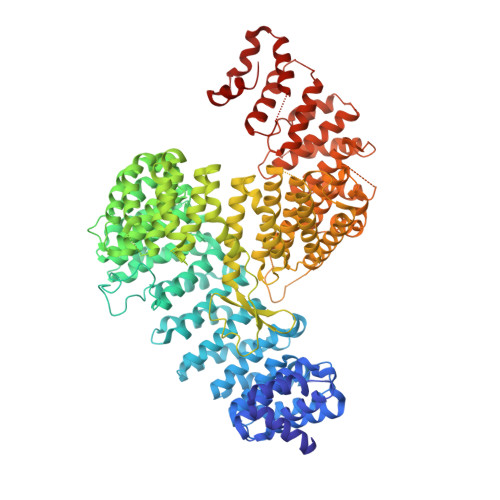
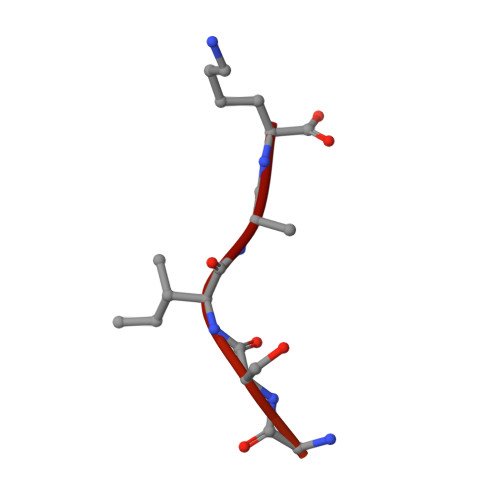
Crystal structure of the karyopherin Kap121p bound to the extreme C-terminus of the protein phosphatase Cdc14p
Kobayashi, J., Hirano, H., Matsuura, Y.(2015) Biochem Biophys Res Commun 463: 309-314
- PubMed: 26022122
- DOI: https://doi.org/10.1016/j.bbrc.2015.05.060
- Primary Citation of Related Structures:
4ZJ7 - PubMed Abstract:
In Saccharomyces cerevisiae, the protein phosphatase Cdc14p is an antagonist of mitotic cyclin-dependent kinases and is a key regulator of late mitotic events such as chromosome segregation, spindle disassembly and cytokinesis. The activity of Cdc14p is controlled by cell-cycle dependent changes in its association with its competitive inhibitor Net1p (also known as Cfi1p) in the nucleolus. For most of the cell cycle up to metaphase, Cdc14p is sequestered in the nucleolus in an inactive state. During anaphase, Cdc14p is released from Net1p, spreads into the nucleus and cytoplasm, and dephosphorylates key mitotic targets. Although regulated nucleocytoplasmic shuttling of Cdc14p has been suggested to be important for exit from mitosis, the mechanism underlying Cdc14p nuclear trafficking remains poorly understood. Here we show that the C-terminal region (residues 517-551) of Cdc14p can function as a nuclear localization signal (NLS) in vivo and also binds to Kap121p (also known as Pse1p), an essential nuclear import carrier in yeast, in a Gsp1p-GTP-dependent manner in vitro. Moreover we report a crystal structure, at 2.4 Å resolution, of Kap121p bound to the C-terminal region of Cdc14p. The structure and structure-based mutational analyses suggest that either the last five residues at the extreme C-terminus of Cdc14p (residues 547-551; Gly-Ser-Ile-Lys-Lys) or adjacent residues with similar sequence (residues 540-544; Gly-Gly-Ile-Arg-Lys) can bind to the NLS-binding site of Kap121p, with two residues (Ile in the middle and Lys at the end of the five residues) of Cdc14p making key contributions to the binding specificity. Based on comparison with other structures of Kap121p-ligand complexes, we propose "IK-NLS" as an appropriate term to refer to the Kap121p-specific NLS.
Organizational Affiliation:
Division of Biological Science, Graduate School of Science, Nagoya University, Japan.