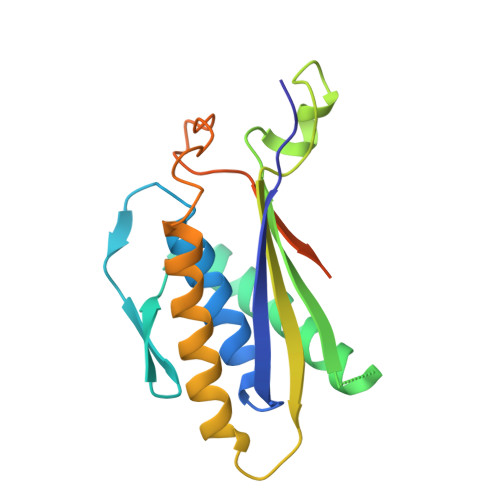
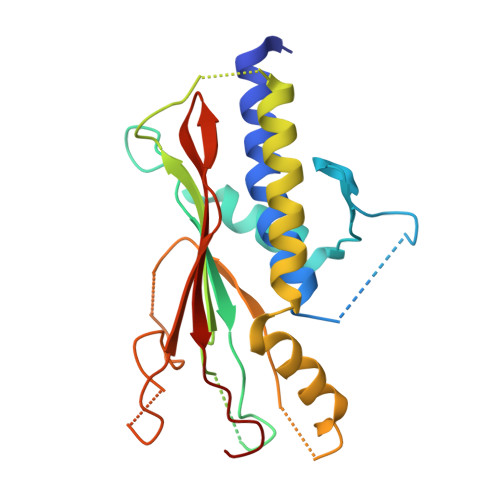
Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation.
Suzuki, H., Kaizuka, T., Mizushima, N., Noda, N.N.(2015) Nat Struct Mol Biol 22: 572-580
- PubMed: 26030876
- DOI: https://doi.org/10.1038/nsmb.3036
- Primary Citation of Related Structures:
4YK8 - PubMed Abstract:
Atg101 is an essential component of the autophagy-initiating ULK complex in higher eukaryotes, but it is absent from the functionally equivalent Atg1 complex in budding yeast. Here, we report the crystal structure of the fission yeast Atg101-Atg13 complex. Atg101 has a Hop1, Rev7 and Mad2 (HORMA) architecture similar to that of Atg13. Mad2 HORMA has two distinct conformations (O-Mad2 and C-Mad2), and, intriguingly, Atg101 resembles O-Mad2 rather than the C-Mad2-like Atg13. Atg13 HORMA from higher eukaryotes possesses an inherently unstable fold, which is stabilized by Atg101 via interactions analogous to those between O-Mad2 and C-Mad2. Mutational studies revealed that Atg101 is responsible for recruiting downstream factors to the autophagosome-formation site in mammals via a newly identified WF finger. These data define the molecular functions of Atg101, providing a basis for elucidating the molecular mechanisms of mammalian autophagy initiation by the ULK complex.
Organizational Affiliation:
Institute of Microbial Chemistry (BIKAKEN), Tokyo, Japan.