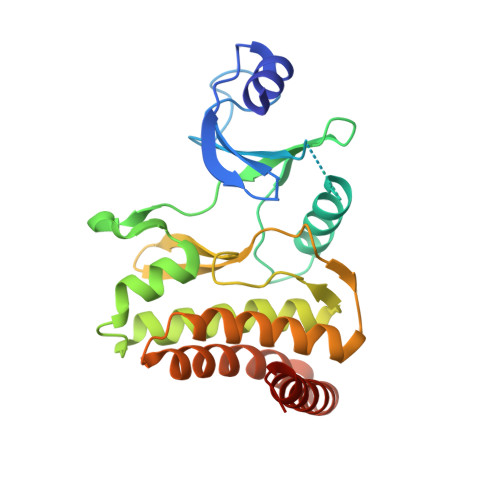
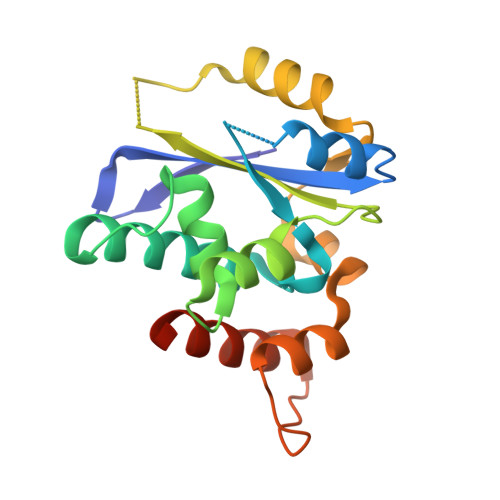
Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Zhang, W., Collinet, B., Graille, M., Daugeron, M.C., Lazar, N., Libri, D., Durand, D., van Tilbeurgh, H.(2015) Nucleic Acids Res 43: 3358-3372
- PubMed: 25735745
- DOI: https://doi.org/10.1093/nar/gkv155
- Primary Citation of Related Structures:
4WW5, 4WW7, 4WW9, 4WWA, 4WX8, 4WXA, 4XAH - PubMed Abstract:
The yeast KEOPS protein complex comprising Kae1, Bud32, Cgi121, Pcc1 and Gon7 is responsible for the essential tRNA threonylcarbamoyladenosine (t(6)A) modification. Deletion of genes coding for the KEOPS subunits also affects telomere elongation and transcriptional regulation. In the present work, the crystal structure of Bud32/Cgi121 in complex with ADP revealed that ADP is bound in the catalytic site of Bud32 in a canonical manner characteristic of Protein Kinase A (PKA) family proteins. We found that Gon7 forms a stable heterodimer with Pcc1 and report the crystal structure of the Pcc1-Gon7 heterodimer. Gon7 interacts with the same Pcc1 region engaged in the archaeal Pcc1 homodimer. We further show that yeast KEOPS, unlike its archaeal counterpart, exists as a heteropentamer in which Gon7, Pcc1, Kae1, Bud32 and Cgi121 also adopt a linear arrangement. We constructed a model of yeast KEOPS that provides structural insight into the role of Gon7. The model also revealed the presence of a highly positively charged crater surrounding the entrance of Kae1 that likely binds tRNA.
Organizational Affiliation:
Institut de Biologie Intégrative de la Cellule, UMR 9198, CNRS, Université de Paris Sud XI, Bâtiment 430, 91405 Orsay, France.