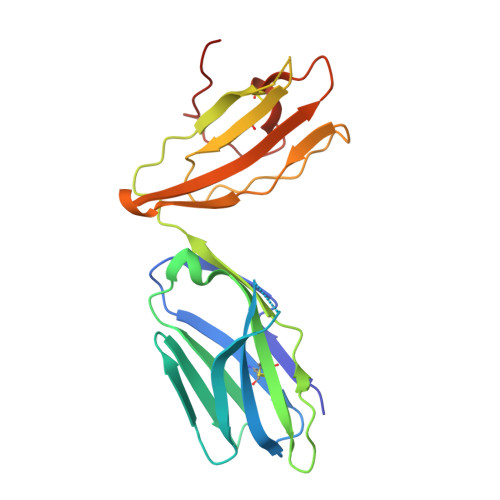
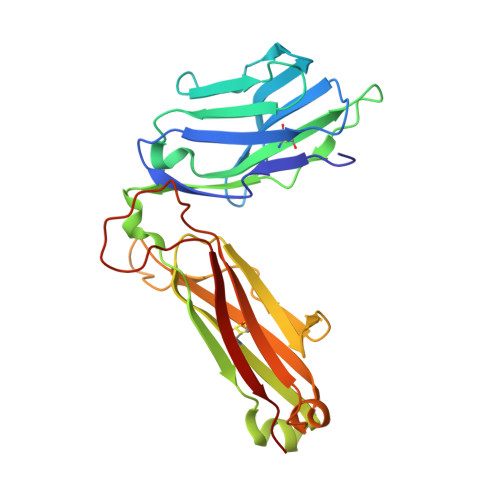
Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Le Nours, J., Praveena, T., Pellicci, D.G., Gherardin, N.A., Ross, F.J., Lim, R.T., Besra, G.S., Keshipeddy, S., Richardson, S.K., Howell, A.R., Gras, S., Godfrey, D.I., Rossjohn, J., Uldrich, A.P.(2016) Nat Commun 7: 10570-10570
- PubMed: 26875526
- DOI: https://doi.org/10.1038/ncomms10570
- Primary Citation of Related Structures:
4WW1, 4WW2, 4WWK - PubMed Abstract:
Crucial to Natural Killer T (NKT) cell function is the interaction between their T-cell receptor (TCR) and CD1d-antigen complex. However, the diversity of the NKT cell repertoire and the ensuing interactions with CD1d-antigen remain unclear. We describe an atypical population of CD1d-α-galactosylceramide (α-GalCer)-reactive human NKT cells that differ markedly from the prototypical TRAV10-TRAJ18-TRBV25-1(+) type I NKT cell repertoire. These cells express a range of TCR α- and β-chains that show differential recognition of glycolipid antigens. Two atypical NKT TCRs (TRAV21-TRAJ8-TRBV7-8 and TRAV12-3-TRAJ27-TRBV6-5) bind orthogonally over the A'-pocket of CD1d, adopting distinct docking modes that contrast with the docking mode of all type I NKT TCR-CD1d-antigen complexes. Moreover, the interactions with α-GalCer differ between the type I and these atypical NKT TCRs. Accordingly, diverse NKT TCR repertoire usage manifests in varied docking strategies and specificities towards CD1d-α-GalCer and related antigens, thus providing far greater scope for diverse glycolipid antigen recognition.
Organizational Affiliation:
Infection and Immunity Program &Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia.