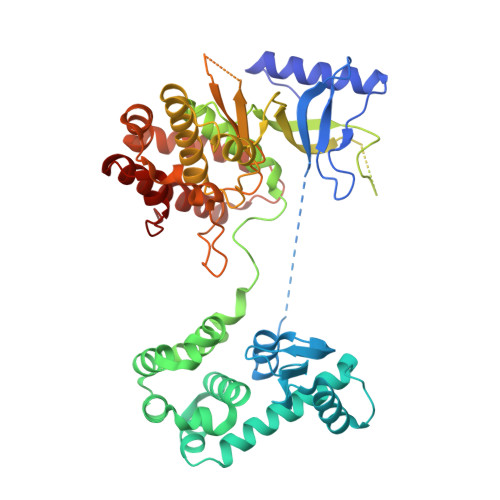
The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Klima, M., Baumlova, A., Chalupska, D., Hrebabecky, H., Dejmek, M., Nencka, R., Boura, E.(2015) Acta Crystallogr D Biol Crystallogr 71: 1555-1563
- PubMed: 26143926
- DOI: https://doi.org/10.1107/S1399004715009505
- Primary Citation of Related Structures:
4WTV, 4YC4 - PubMed Abstract:
Phosphatidylinositol 4-phosphate (PI4P) is the most abundant monophosphoinositide in eukaryotic cells. Humans have four phosphatidylinositol 4-kinases (PI4Ks) that synthesize PI4P, among which are PI4K IIβ and PI4K IIα. In this study, two crystal structures are presented: the structure of human PI4K IIβ and the structure of PI4K IIα containing a nucleoside analogue. The former, a complex with ATP, is the first high-resolution (1.9 Å) structure of a PI4K. These structures reveal new details such as high conformational heterogeneity of the lateral hydrophobic pocket of the C-lobe and together provide a structural basis for isoform-specific inhibitor design.
Organizational Affiliation:
Department of Biochemistry, Institute of Organic Chemistry and Biochemistry, Flemingovo nam. 2, 166 10 Prague, Czech Republic.