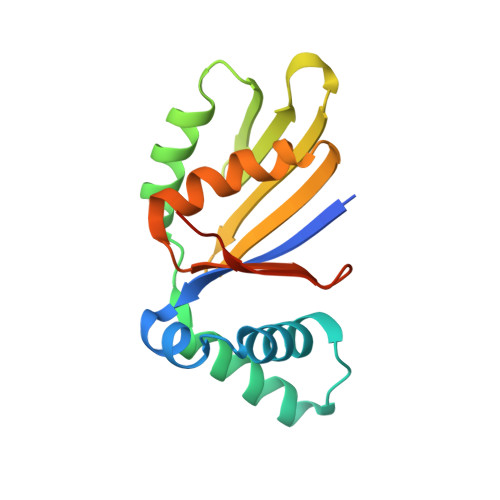
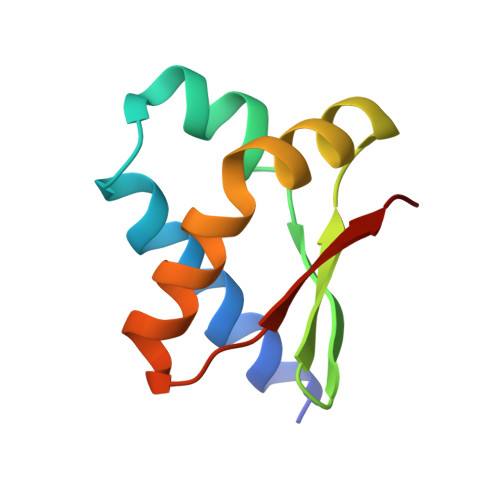
Structure and interactions of the Bacillus subtilis sporulation inhibitor of DNA replication, SirA, with domain I of DnaA.
Jameson, K.H., Rostami, N., Fogg, M.J., Turkenburg, J.P., Grahl, A., Murray, H., Wilkinson, A.J.(2014) Mol Microbiol 93: 975-991
- PubMed: 25041308
- DOI: https://doi.org/10.1111/mmi.12713
- Primary Citation of Related Structures:
4TPS - PubMed Abstract:
Chromosome copy number in cells is controlled so that the frequency of initiation of DNA replication matches that of cell division. In bacteria, this is achieved through regulation of the interaction between the initiator protein DnaA and specific DNA elements arrayed at the origin of replication. DnaA assembles at the origin and promotes DNA unwinding and the assembly of a replication initiation complex. SirA is a DnaA-interacting protein that inhibits initiation of replication in diploid Bacillus subtilis cells committed to the developmental pathway leading to formation of a dormant spore. Here we present the crystal structure of SirA in complex with the N-terminal domain of DnaA revealing a heterodimeric complex. The interacting surfaces of both proteins are α-helical with predominantly apolar side-chains packing in a hydrophobic interface. Site-directed mutagenesis experiments confirm the importance of this interface for the interaction of the two proteins in vitro and in vivo. Localization of GFP-SirA indicates that the protein accumulates at the replisome in sporulating cells, likely through a direct interaction with DnaA. The SirA interacting surface of DnaA corresponds closely to the HobA-interacting surface of DnaA from Helicobacter pylori even though HobA is an activator of DnaA and SirA is an inhibitor.
Organizational Affiliation:
Structural Biology Laboratory, Department of Chemistry, University of York, York, YO10 5DD, UK.