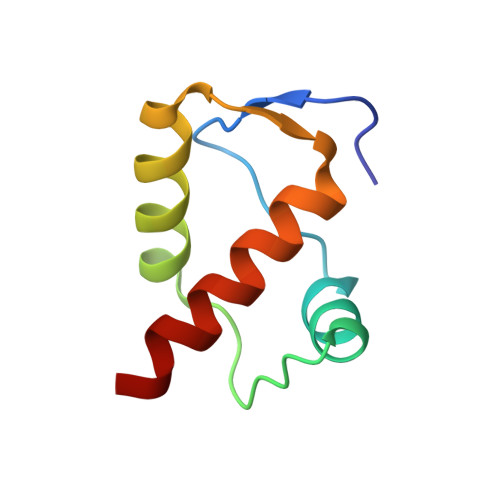
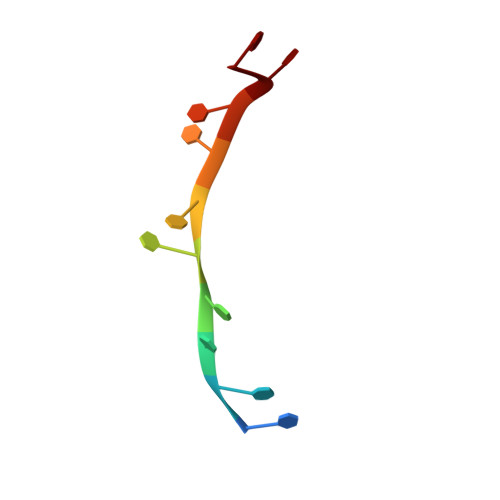
Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode.
Liu, Z., Chen, P., Wang, X., Cai, G., Niu, L., Teng, M., Li, X.(2014) Nucleic Acids Res 42: 9470-9483
- PubMed: 25053836
- DOI: https://doi.org/10.1093/nar/gku633
- Primary Citation of Related Structures:
4OU6, 4OU7 - PubMed Abstract:
DnaT is a primosomal protein that is required for the stalled replication fork restart in Escherichia coli. As an adapter, DnaT mediates the PriA-PriB-ssDNA ternary complex and the DnaB/C complex. However, the fundamental function of DnaT during PriA-dependent primosome assembly is still a black box. Here, we report the 2.83 Å DnaT(84-153)-dT10 ssDNA complex structure, which reveals a novel three-helix bundle single-stranded DNA binding mode. Based on binding assays and negative-staining electron microscopy results, we found that DnaT can bind to phiX 174 ssDNA to form nucleoprotein filaments for the first time, which indicates that DnaT might function as a scaffold protein during the PriA-dependent primosome assembly. In combination with biochemical analysis, we propose a cooperative mechanism for the binding of DnaT to ssDNA and a possible model for the assembly of PriA-PriB-ssDNA-DnaT complex that sheds light on the function of DnaT during the primosome assembly and stalled replication fork restart. This report presents the first structure of the DnaT C-terminal complex with ssDNA and a novel model that explains the interactions between the three-helix bundle and ssDNA.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China Key Laboratory of Structural Biology, Chinese Academy of Sciences, Hefei, Anhui 230026, People's Republic of China.