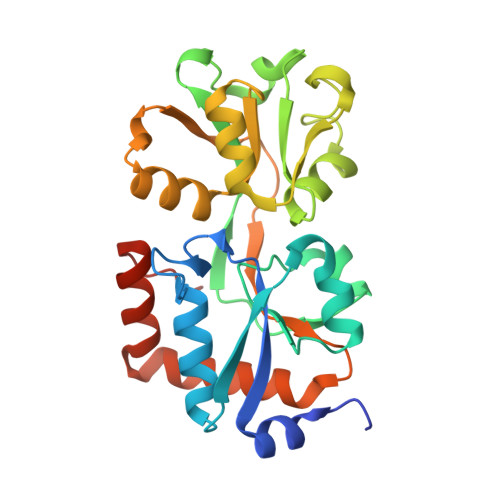
Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
Brunzelle, J.S., Wawrzak, W., Yim, Y., Kudritska, M., Savchenko, A., Anderson, W.F., Center for Structural Genomics of Infectious DiseasesTo be published.