Structure of a binary complex between homologous tetrameric legume lectins from Butea monosperma and Spatholobus parviflorus seeds
Surya, S., Abhilash, J., Geethanandan, K., Sadasivan, C., Haridas, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lectin Alpha chain | 255 | Butea monosperma | Mutation(s): 0 | 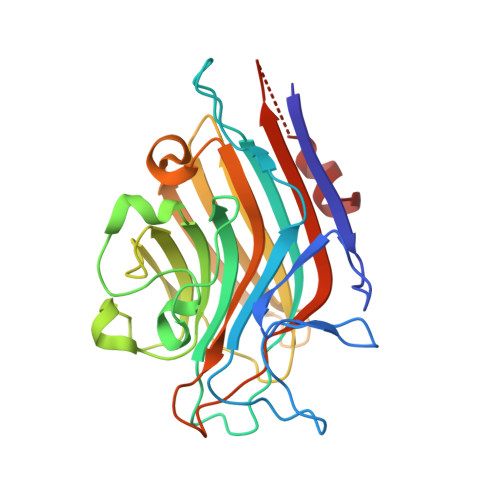 | |
UniProt | |||||
Find proteins for H2L2M6 (Butea monosperma) Explore H2L2M6 Go to UniProtKB: H2L2M6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H2L2M6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lectin Beta Chain | 239 | Butea monosperma | Mutation(s): 0 | 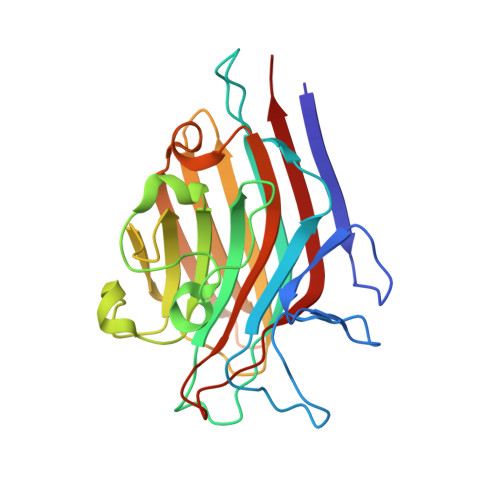 | |
UniProt | |||||
Find proteins for H2L2M7 (Butea monosperma) Explore H2L2M7 Go to UniProtKB: H2L2M7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H2L2M7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Seed lectin alpha chain | 251 | Spatholobus parviflorus | Mutation(s): 0 | 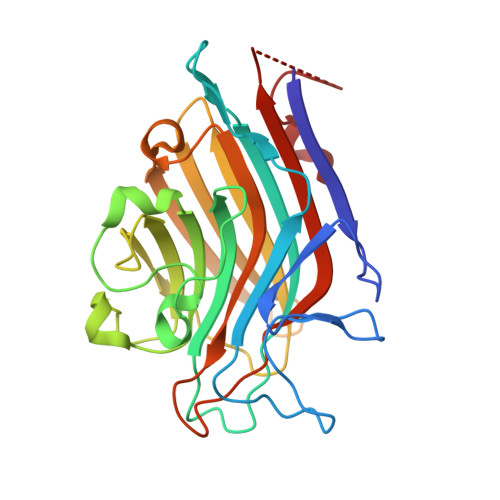 | |
UniProt | |||||
Find proteins for P86352 (Spatholobus parviflorus) Explore P86352 Go to UniProtKB: P86352 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P86352 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Seed lectin beta chain | 239 | Spatholobus parviflorus | Mutation(s): 0 | 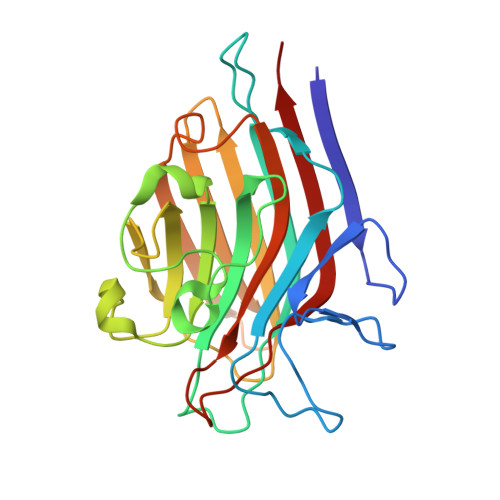 | |
UniProt | |||||
Find proteins for P86353 (Spatholobus parviflorus) Explore P86353 Go to UniProtKB: P86353 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P86353 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ABU Query on ABU | GA [auth H] K [auth A] N [auth B] Q [auth C] U [auth D] | GAMMA-AMINO-BUTANOIC ACID C4 H9 N O2 BTCSSZJGUNDROE-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | BA [auth F], R [auth C], V [auth D] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| MN Query on MN | AA [auth F] DA [auth G] FA [auth H] J [auth A] M [auth B] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| CA Query on CA | CA [auth G] EA [auth H] I [auth A] L [auth B] O [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 77.11 | α = 96 |
| b = 78.63 | β = 89.99 |
| c = 96.19 | γ = 100.37 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| Auto | model building |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| Auto | phasing |