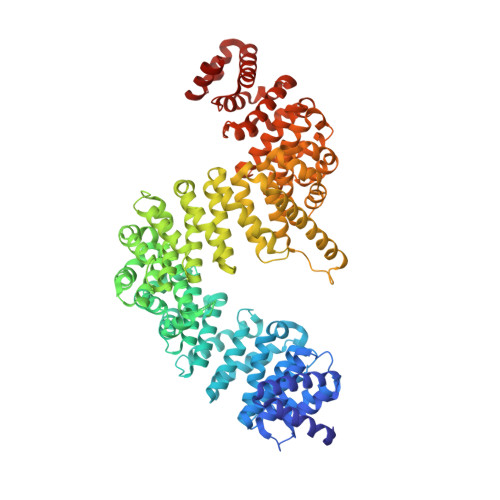
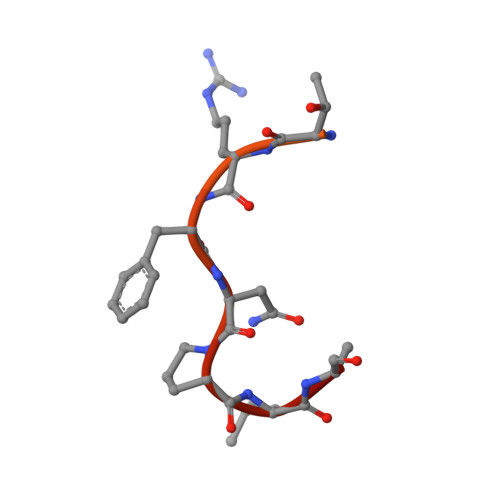
Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
Soniat, M., Sampathkumar, P., Collett, G., Gizzi, A.S., Banu, R.N., Bhosle, R.C., Chamala, S., Chowdhury, S., Fiser, A., Glenn, A.S., Hammonds, J., Hillerich, B., Khafizov, K., Love, J.D., Matikainen, B., Seidel, R.D., Toro, R., Rajesh Kumar, P., Bonanno, J.B., Chook, Y.M., Almo, S.C.(2013) J Struct Funct Genomics 14: 31-35
- PubMed: 23535894
- DOI: https://doi.org/10.1007/s10969-013-9150-1
- Primary Citation of Related Structures:
4JLQ - PubMed Abstract:
Import-Karyopherin or Importin proteins bind nuclear localization signals (NLSs) to mediate the import of proteins into the cell nucleus. Karyopherin β2 or Kapβ2, also known as Transportin, is a member of this transporter family responsible for the import of numerous RNA binding proteins. Kapβ2 recognizes a targeting signal termed the PY-NLS that lies within its cargos to target them through the nuclear pore complex. The recognition of PY-NLS by Kapβ2 is conserved throughout eukaryotes. Kap104, the Kapβ2 homolog in Saccharomyces cerevisiae, recognizes PY-NLSs in cargos Nab2, Hrp1, and Tfg2. We have determined the crystal structure of Kapβ2 bound to the PY-NLS of the mRNA processing protein Nab2 at 3.05-Å resolution. A seven-residue segment of the PY-NLS of Nab2 is observed to bind Kapβ2 in an extended conformation and occupies the same PY-NLS binding site observed in other Kapβ2·PY-NLS structures.
Organizational Affiliation:
Department of Pharmacology, University of Texas Southwestern, Dallas, TX 75390, USA. michael.soniat@utsouthwestern.edu