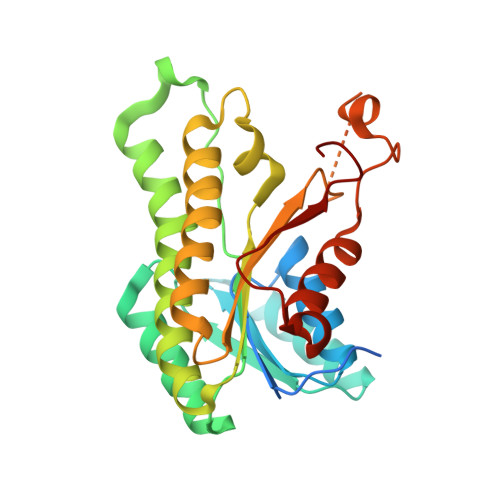
Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Helicobacter pylori 26695 complexed with NAD+
Hou, J., Osinski, T., Zheng, H., Shumilin, I., Shabalin, I., Shatsman, S., Anderson, W.F., Minor, W., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.