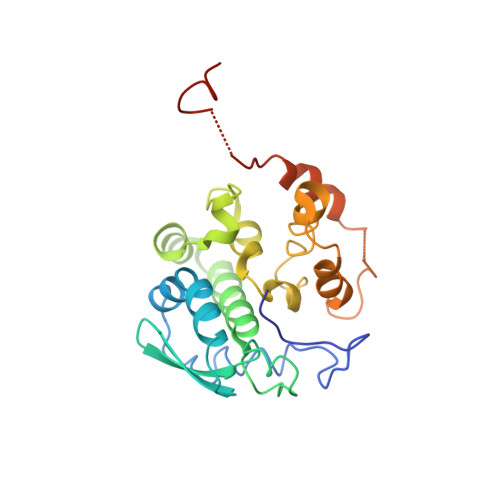
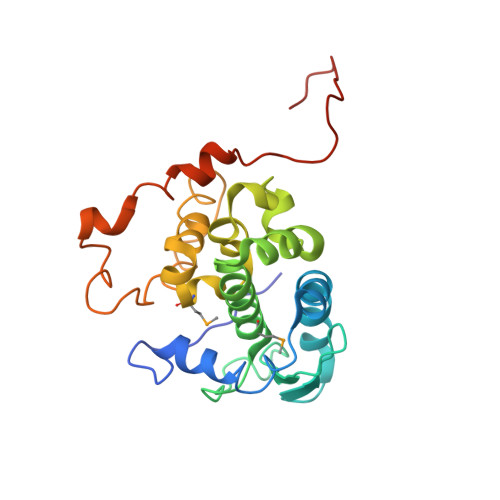
Structure of Schmallenberg orthobunyavirus nucleoprotein suggests a novel mechanism of genome encapsidation
Dong, H., Li, P., Elliott, R.M., Dong, C.(2013) J Virol 87: 5593-5601
- PubMed: 23468499
- DOI: https://doi.org/10.1128/JVI.00223-13
- Primary Citation of Related Structures:
4IDU, 4IDX - PubMed Abstract:
Schmallenberg virus (SBV), a newly emerged orthobunyavirus (family Bunyaviridae), has spread rapidly across Europe and has caused congenital abnormalities in the offspring of cattle, sheep, and goats. Like other orthobunyaviruses, SBV contains a tripartite negative-sense RNA genome that encodes four structural and two nonstructural proteins. The nucleoprotein (N) encapsidates the three viral genomic RNA segments and plays a crucial role in viral RNA transcription and replication. Here we report the crystal structure of the bacterially expressed SBV nucleoprotein to a 3.06-Å resolution. The protomer is composed of two domains (N-terminal and C-terminal domains) with flexible N-terminal and C-terminal arms. The N protein has a novel fold and forms a central positively charged cleft for genomic RNA binding. The nucleoprotein purified under native conditions forms a tetramer, while the nucleoprotein obtained following denaturation and refolding forms a hexamer. Our structural and functional analyses demonstrate that both N-terminal and C-terminal arms are involved in N-N interaction and oligomerization and play an essential role in viral RNA synthesis, suggesting a novel mechanism for viral RNA encapsidation and transcription.
Organizational Affiliation:
Biomedical Research Centre, Norwich Medical School, University of East Anglia, Norwich Research Park, Norwich, United Kingdom.