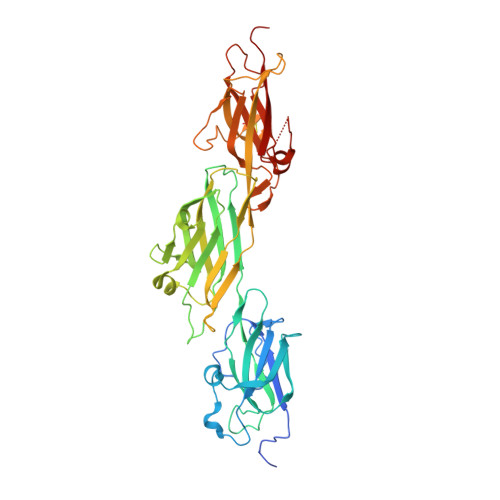
A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly.
Kang, H.J., Paterson, N.G., Kim, C.U., Middleditch, M., Chang, C., Ton-That, H., Baker, E.N.(2014) Acta Crystallogr D Biol Crystallogr 70: 1190-1201
- PubMed: 24816089
- DOI: https://doi.org/10.1107/S1399004714001400
- Primary Citation of Related Structures:
4HSQ, 4HSS - PubMed Abstract:
The Gram-positive organism Corynebacterium diphtheriae, the cause of diphtheria in humans, expresses pili on its surface which it uses for adhesion and colonization of its host. These pili are covalent protein polymers composed of three types of pilin subunit that are assembled by specific sortase enzymes. A structural analysis of the major pilin SpaD, which forms the polymeric backbone of one of the three types of pilus expressed by C. diphtheriae, is reported. Mass-spectral and crystallographic analysis shows that SpaD contains three internal Lys-Asn isopeptide bonds. One of these, shown by mass spectrometry to be located in the N-terminal D1 domain of the protein, only forms slowly, implying an energy barrier to bond formation. Two crystal structures, of the full-length three-domain protein at 2.5 Å resolution and of a two-domain (D2-D3) construct at 1.87 Å resolution, show that each of the three Ig-like domains contains a single Lys-Asn isopeptide-bond cross-link, assumed to give mechanical stability as in other such pili. Additional stabilizing features include a disulfide bond in the D3 domain and a calcium-binding loop in D2. The N-terminal D1 domain is more flexible than the others and, by analogy with other major pilins of this type, the slow formation of its isopeptide bond can be attributed to its location adjacent to the lysine used in sortase-mediated polymerization during pilus assembly.
Organizational Affiliation:
Maurice Wilkins Centre for Molecular Biodiscovery and School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland 1142, New Zealand.