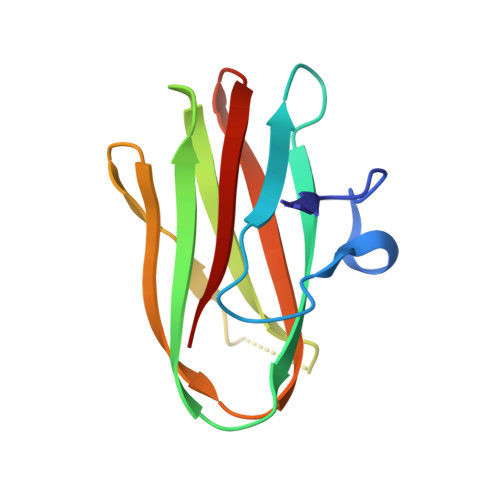
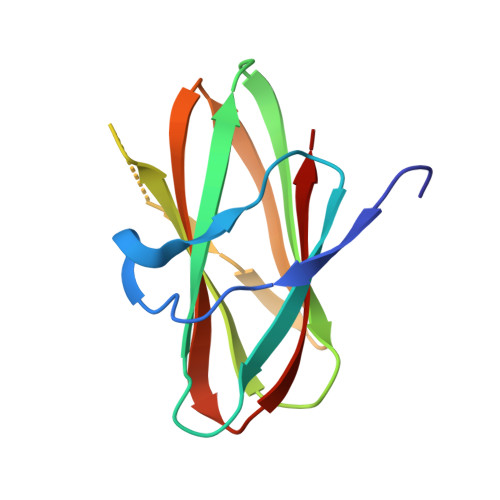
Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
Bauer, R., Wilson, J.J., Philominathan, S.T., Davis, D., Matsushita, O., Sakon, J.(2013) J Bacteriol 195: 318-327
- PubMed: 23144249
- DOI: https://doi.org/10.1128/JB.00010-12
- Primary Citation of Related Structures:
3JQW, 3JQX, 4HPK - PubMed Abstract:
Clostridium histolyticum secretes collagenases, ColG and ColH, that cause extensive tissue destruction in myonecrosis. The C-terminal collagen-binding domain (CBD) of collagenase is required for insoluble collagen fibril binding and subsequent collagenolysis. The high-resolution crystal structures of ColG-CBD (s3b) and ColH-CBD (s3) are reported in this paper. The new X-ray structure of s3 was solved at 2.0-Å resolution (R = 17.4%; R(free) = 23.3%), while the resolution of the previously determined s3b was extended to 1.4 Å (R = 17.9%; R(free) = 21.0%). Despite sharing only 30% sequence identity, the molecules resemble one another closely (root mean square deviation [RMSD] C(α) = 1.5 Å). All but one residue, whose side chain chelates with Ca(2+), are conserved. The dual Ca(2+) binding site in s3 is completed by an unconserved aspartate. Differential scanning calorimetric measurements showed that s3 gains thermal stability, comparable to s3b, by binding to Ca(2+) (holo T(m) = 94.1°C; apo T(m) = 70.2°C). holo s3 is also stabilized against chemical denaturants urea and guanidine HCl. The three most critical residues for collagen interaction in s3b are conserved in s3. The general shape of the binding pocket is retained by altered loop structures and side chain positions. Small-angle X-ray scattering data revealed that s3 also binds asymmetrically to minicollagen. Besides the calcium-binding sites and the collagen-binding pocket, architecturally important hydrophobic residues and the hydrogen-bonding network around the cis-peptide bond are well conserved within the metallopeptidase subfamily M9B. CBDs were previously shown to bind to the extracellular matrix of various tissues. Compactness and extreme stability in physiological Ca(2+) concentration possibly make both CBDs suitable for targeted growth factor delivery.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Arkansas, Fayetteville, AR, USA.