The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
Neubauer, J.L., Pham, T., Immormino, R.M., Dollins, D.E., Endo-Streeter, S.T., Li, S., Pemble IV, C.W., York, J.D.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-terminal acetyltransferase A complex subunit NAT1 | 863 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: NAT1, AAA1, YDL040C, D2720 | 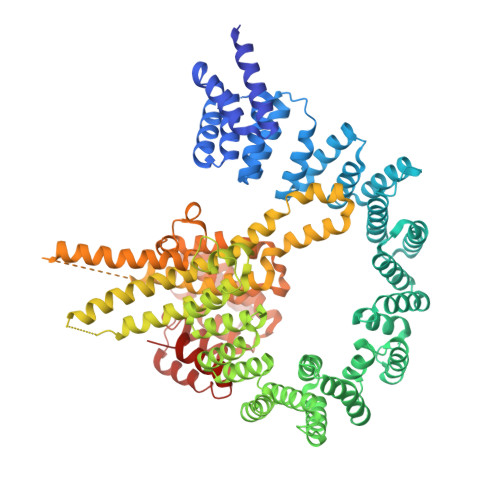 | |
UniProt | |||||
Find proteins for P12945 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P12945 Go to UniProtKB: P12945 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12945 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N-terminal acetyltransferase A complex catalytic subunit ARD1 | 248 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: ARD1, YHR013C EC: 2.3.1.88 | 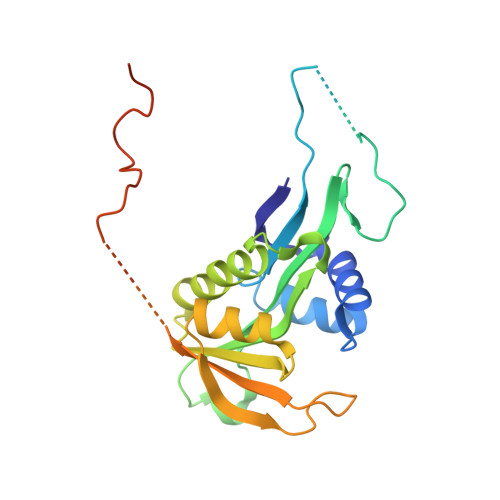 | |
UniProt | |||||
Find proteins for P07347 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P07347 Go to UniProtKB: P07347 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07347 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IHP Query on IHP | C [auth A] | INOSITOL HEXAKISPHOSPHATE C6 H18 O24 P6 IMQLKJBTEOYOSI-GPIVLXJGSA-N |  | ||
| PG4 Query on PG4 | D [auth A], E [auth A], F [auth A], G [auth A], H [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 135.662 | α = 90 |
| b = 135.662 | β = 90 |
| c = 175.801 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| CNS | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |