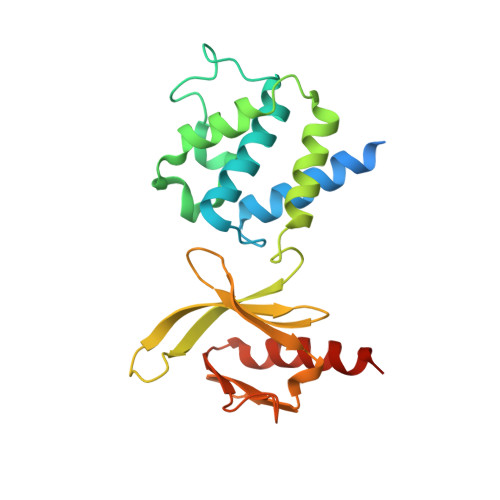
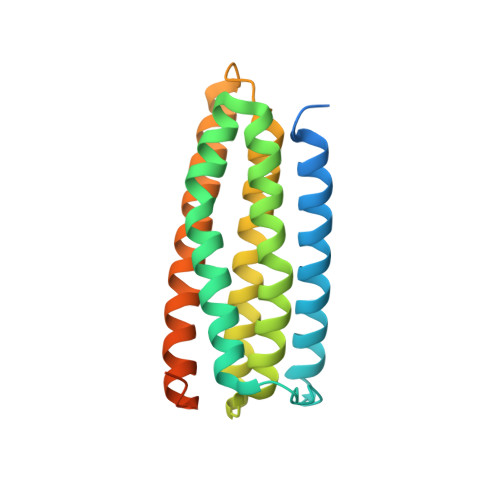
A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion.
Song, X., Yang, J., Hirbawi, J., Ye, S., Perera, H.D., Goksoy, E., Dwivedi, P., Plow, E.F., Zhang, R., Qin, J.(2012) Cell Res 22: 1533-1545
- PubMed: 22710802
- DOI: https://doi.org/10.1038/cr.2012.97
- Primary Citation of Related Structures:
4F7G - PubMed Abstract:
The activation of heterodimeric (α/β) integrin transmembrane receptors by cytosolic protein talin is crucial for regulating diverse cell-adhesion-dependent processes, including blood coagulation, tissue remodeling, and cancer metastasis. This process is triggered by the coincident binding of N-terminal FERM (four-point-one-protein/ezrin/radixin/moesin) domain of talin (talin-FERM) to the inner membrane surface and integrin β cytoplasmic tail, but how these binding events are spatiotemporally regulated remains obscure. Here we report the crystal structure of a dormant talin, revealing how a C-terminal talin rod segment (talin-RS) self-masks a key integrin-binding site on talin-FERM via a large interface. Unexpectedly, the structure also reveals a distinct negatively charged surface on talin-RS that electrostatically hinders the talin-FERM binding to the membrane. Such a dual inhibitory topology for talin is consistent with the biochemical and functional data, but differs significantly from a previous model. We show that upon enrichment with phosphotidylinositol-4,5-bisphosphate (PIP2) - a known talin activator, membrane strongly attracts a positively charged surface on talin-FERM and simultaneously repels the negatively charged surface on talin-RS. Such an electrostatic "pull-push" process promotes the relief of the dual inhibition of talin-FERM, which differs from the classic "steric clash" model for conventional PIP2-induced FERM domain activation. These data therefore unravel a new type of membrane-dependent FERM domain regulation and illustrate how it mediates the talin on/off switches to regulate integrin transmembrane signaling and cell adhesion.
Organizational Affiliation:
Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.