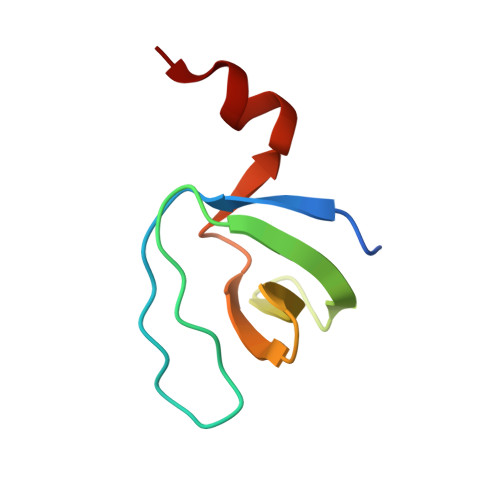
Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Liu, X., Chen, M., Lobo, P., An, J., Grace Cheng, S.W., Moradian, A., Morin, G.B., Van Petegem, F., Jiang, X.(2012) Proteomics 12: 2094-2106
- PubMed: 22623184
- DOI: https://doi.org/10.1002/pmic.201100553
- Primary Citation of Related Structures:
4ESR - PubMed Abstract:
ABL tyrosine kinase inhibitor (TKI) therapy induces clinical remission in chronic myeloid leukemia (CML) patients but early relapses and later emergence of TKI-resistant disease remain problematic. We recently demonstrated that the AHI-1 oncogene physically interacts with BCR-ABL and JAK2 and mediates cellular resistance to TKI in CML stem/progenitor cells. We now show that deletion of the SH3 domain of AHI-1 significantly enhances apoptotic response of BCR-ABL(+) cells to TKIs compared to cells expressing full-length AHI-1. We have also discovered a novel interaction between AHI-1 and Dynamin-2, a GTPase, through the AHI-1 SH3 domain. The crystal structure of the AHI-1 SH3 domain at 1.53-Å resolution reveals that it adopts canonical SH3 folding, with the exception of an unusual C-terminal α helix. PD1R peptide, known to interact with the PI3K SH3 domain, was used to model the binding pattern between the AHI-1 SH3 domain and its ligands. These studies showed that an "Arg-Arg-Trp" stack may form within the binding interface, providing a potential target site for designing specific drugs. The crystal structure of the AHI-1 SH3 domain thus provides a valuable tool for identification of key interaction sites in regulation of drug resistance and for the development of small molecule inhibitors for CML.
Organizational Affiliation:
Terry Fox Laboratory, British Columbia Cancer Agency, Vancouver, BC, Canada.