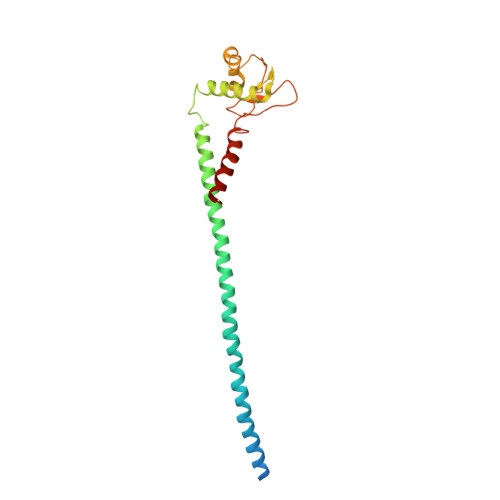
The structure of subunit E of the Pyrococcus horikoshii OT3 A-ATP synthase gives insight into the elasticity of the peripheral stalk.
Balakrishna, A.M., Hunke, C., Gruber, G.(2012) J Mol Biol 420: 155-163
- PubMed: 22516614
- DOI: https://doi.org/10.1016/j.jmb.2012.04.012
- Primary Citation of Related Structures:
4DT0 - PubMed Abstract:
A(1)A(O) ATP synthases are the major energy converters of archaea. They are composed of an A(1) region that synthesizes ATP and an integral part A(O) that conducts ions. Subunit E is a component of the peripheral stalk that links the A(1) with the A(O) part of the A-ATP synthase. We have determined the crystal structure of the entire subunit E (PhE) of the Pyrococcus horikoshii OT3 A-ATP synthase at 3.6 Å resolution. The structure reveals an extended S-shaped N-terminal α-helix with 112.29 Å in length, followed by a globular head group. The S-shaped feature, common in elastic connectors and spacers, would facilitate the storage of transient elastic energy during rotary motion in the enzyme. The structure has been superimposed into the asymmetric peripheral stalks of the three-dimensional reconstruction of the Pyrococcus furiosus enzyme, revealing that the S-shaped subunit PhE fits well into the bent peripheral stalk, whereas the previously solved E subunit structure (3.1 Å resolution) of Thermus thermophilus A-ATP synthase is well accommodated in the density of the straight stator domain. The different features of the two stalk subunits are discussed in light of a novel coupling mechanism in A-ATP synthases proposed to differ from the Wankel engine of F-ATP synthases.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Republic of Singapore.