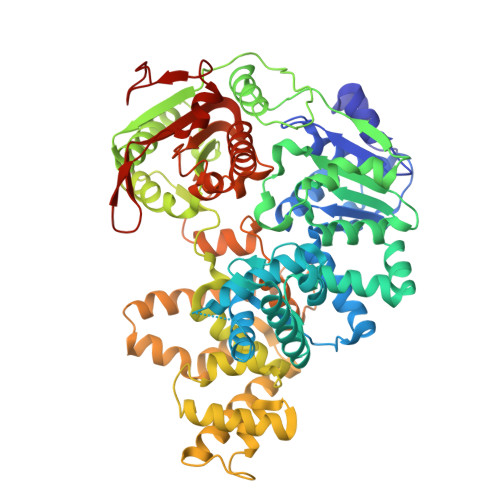
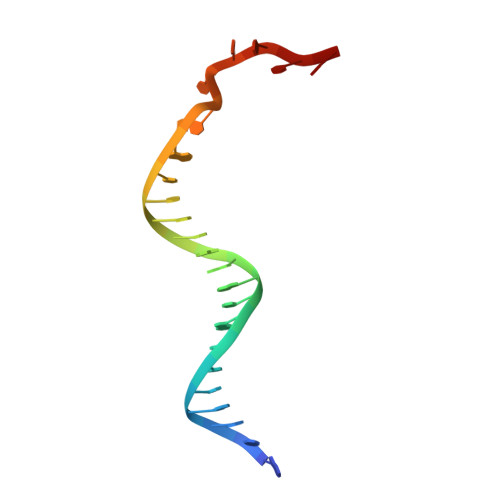
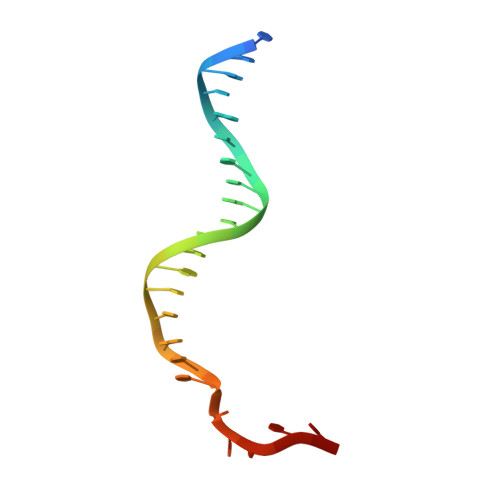
Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Stelter, M., Acajjaoui, S., Mcsweeney, S., Timmins, J.(2013) PLoS One 8: 77364
- PubMed: 24143224
- DOI: https://doi.org/10.1371/journal.pone.0077364
- Primary Citation of Related Structures:
4C2T, 4C2U, 4C30 - PubMed Abstract:
DNA helicases are responsible for unwinding the duplex DNA, a key step in many biological processes. UvrD is a DNA helicase involved in several DNA repair pathways. We report here crystal structures of Deinococcus radiodurans UvrD (drUvrD) in complex with DNA in different nucleotide-free and bound states. These structures provide us with three distinct snapshots of drUvrD in action and for the first time trap a DNA helicase undergoing a large-scale spiral movement around duplexed DNA. Our structural data also improve our understanding of the molecular mechanisms that regulate DNA unwinding by Superfamily 1A (SF1A) helicases. Our biochemical data reveal that drUvrD is a DNA-stimulated ATPase, can translocate along ssDNA in the 3'-5' direction and shows ATP-dependent 3'-5', and surprisingly also, 5'-3' helicase activity. Interestingly, we find that these translocase and helicase activities of drUvrD are modulated by the ssDNA binding protein. Analysis of drUvrD mutants indicate that the conserved β-hairpin structure of drUvrD that functions as a separation pin is critical for both drUvrD's 3'-5' and 5'-3' helicase activities, whereas the GIG motif of drUvrD involved in binding to the DNA duplex is essential for the 5'-3' helicase activity only. These special features of drUvrD may reflect its involvement in a wide range of DNA repair processes in vivo.
Organizational Affiliation:
Structural Biology Group, European Synchrotron Radiation Facility, Grenoble, France ; University Grenoble Alpes, Institut de Biologie structurale, Grenoble, France ; Centre National de la Recherche Scientifique, Institut de Biologie structurale, Grenoble, France ; Commissariat à l'énergie atomique et aux énergies alternatives, Département du Science du Vivant, Institut de Biologie structurale, Grenoble, France.