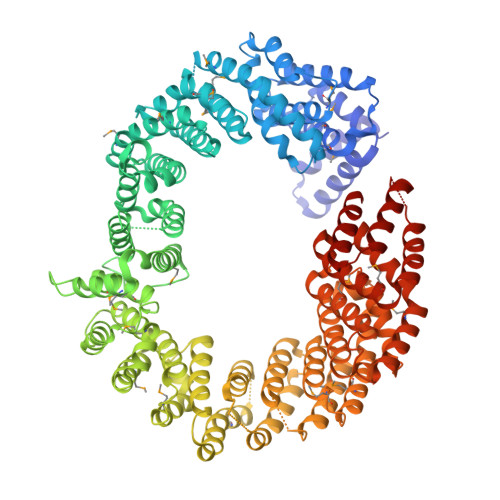
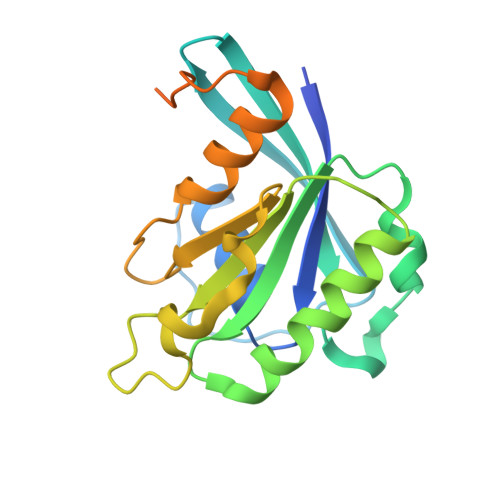
Structural Basis for Nuclear Import of Splicing Factors by Human Transportin 3.
Maertens, G.N., Cook, N.J., Wang, W., Hare, S., Gupta, S.S., Oztop, I., Lee, K., Pye, V.E., Cosnefroy, O., Snijders, A.P., Kewalramani, V.N., Fassati, A., Engelman, A., Cherepanov, P.(2014) Proc Natl Acad Sci U S A 111: 2728
- PubMed: 24449914
- DOI: https://doi.org/10.1073/pnas.1320755111
- Primary Citation of Related Structures:
4C0O, 4C0P, 4C0Q - PubMed Abstract:
Transportin 3 (Tnpo3, Transportin-SR2) is implicated in nuclear import of splicing factors and HIV-1 replication. Herein, we show that the majority of cellular Tnpo3 binding partners contain arginine-serine (RS) repeat domains and present crystal structures of human Tnpo3 in its free as well as GTPase Ran- and alternative splicing factor/splicing factor 2 (ASF/SF2)-bound forms. The flexible β-karyopherin fold of Tnpo3 embraces the RNA recognition motif and RS domains of the cargo. A constellation of charged residues on and around the arginine-rich helix of Tnpo3 HEAT repeat 15 engage the phosphorylated RS domain and are critical for the recognition and nuclear import of ASF/SF2. Mutations in the same region of Tnpo3 impair its interaction with the cleavage and polyadenylation specificity factor 6 (CPSF6) and its ability to support HIV-1 replication. Steric incompatibility of the RS domain and RanGTP engagement by Tnpo3 provides the mechanism for cargo release in the nucleus. Our results elucidate the structural bases for nuclear import of splicing factors and the Tnpo3-CPSF6 nexus in HIV-1 biology.
Organizational Affiliation:
Division of Infectious Diseases, Imperial College London, St. Mary's Campus, London W2 1PG, United Kingdom.