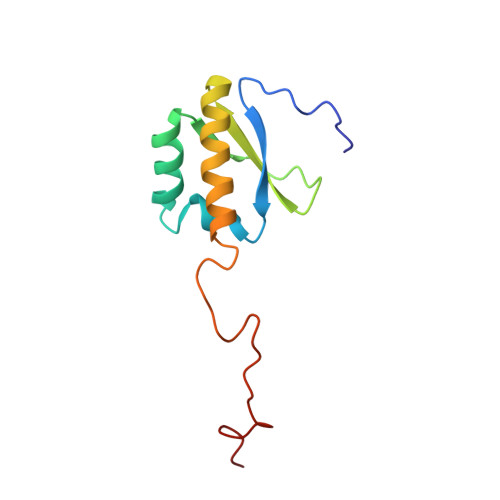
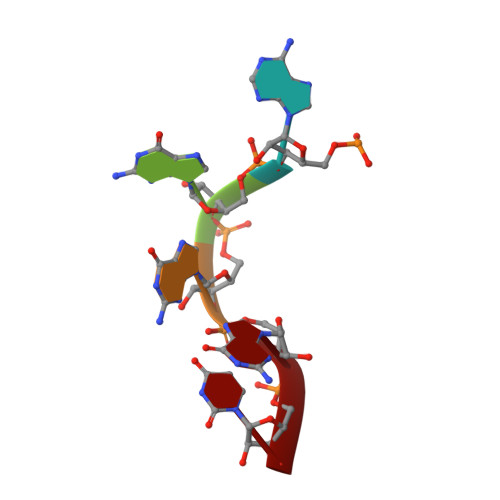
Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nicastro, G., Garcia-Mayoral, M.F., Hollingworth, D., Kelly, G., Martin, S.R., Briata, P., Gherzi, R., Ramos, A.(2012) Nat Struct Mol Biol 19: 1282
- PubMed: 23142982
- DOI: https://doi.org/10.1038/nsmb.2427
- Primary Citation of Related Structures:
4B8T - PubMed Abstract:
Let-7 is an important tumor-suppressive microRNA (miRNA) that acts as an on-off switch for cellular differentiation and regulates the expression of a set of human oncogenes. Binding of the human KSRP protein to let-7 miRNA precursors positively regulates their processing to mature let-7, thereby contributing to control of cell proliferation, apoptosis and differentiation. Here we analyze the molecular basis for KSRP-let-7 precursor selectivity and show how the third KH domain of the protein recognizes a G-rich sequence in the pre-let-7 terminal loop and dominates the interaction. The structure of the KH3-RNA complex explains the protein recognition of this noncanonical KH target sequence, and we demonstrate that the specificity of this binding is crucial for the functional interaction between the protein and the miRNA precursor.
Organizational Affiliation:
Molecular Structure Division, Medical Research Council National Institute for Medical Research, London, UK.