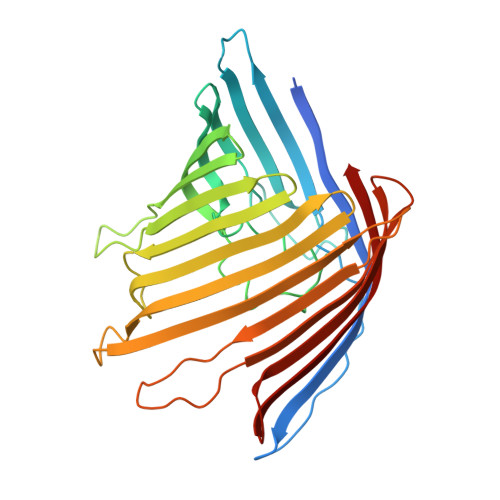
Structure and Function of the Porb Porin from Disseminating Neisseria Gonorrhoeae.
Zeth, K., Kozjak-Pavlovic, V., Faulstich, M., Fraunholz, M., Hurwitz, R., Kepp, O., Rudel, T.(2013) Biochem J 449: 631
- PubMed: 23095086
- DOI: https://doi.org/10.1042/BJ20121025
- Primary Citation of Related Structures:
4AUI - PubMed Abstract:
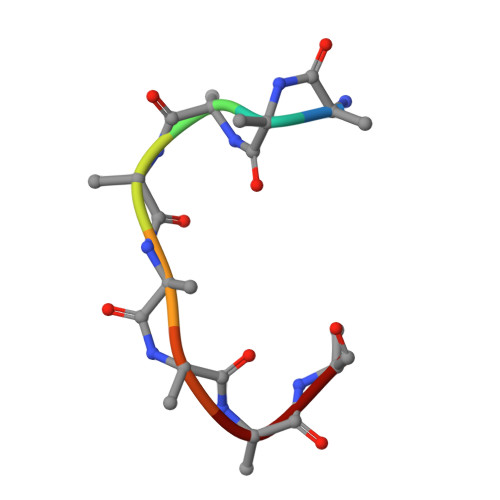
The outer membrane of Gram-negative bacteria contains a large number of channel-forming proteins, porins, for the uptake of small nutrient molecules. Neisseria gonorrhoeae PorBIA (PorB of serotype A) are associated with disseminating diseases and mediate a rapid bacterial invasion into host cells in a phosphate-sensitive manner. To gain insights into this structure-function relationship we analysed PorBIA by X-ray crystallography in the presence of phosphate and ATP. The structure of PorBIA in the complex solved at a resolution of 3.3 Å (1 Å=0.1 nm) displays a surplus of positive charges inside the channel. ATP ligand-binding in the channel is co-ordinated by the positively charged residues of the channel interior. These residues ligate the aromatic, sugar and pyrophosphate moieties of the ligand. Two phosphate ions were observed in the structure, one of which clamped by two arginine residues (Arg92 and Arg124) localized at the extraplasmic channel exit. A short β-bulge in β2-strand together with the long L3 loop narrow the barrel diameter significantly and further support substrate specificity through hydrogen bond interactions. Interestingly the structure also comprised a small peptide as a remnant of a periplasmic protein which physically links porin molecules to the peptidoglycan network. To test the importance of Arg92 on bacterial invasion the residue was mutated. In vivo assays of bacteria carrying a R92S mutation confirmed the importance of this residue for host-cell invasion. Furthermore systematic sequence and structure comparisons of PorBIA from Neisseriaceae indicated Arg92 to be unique in disseminating N. gonorrhoeae thereby possibly distinguishing invasion-promoting porins from other neisserial porins.
Organizational Affiliation:
Max Planck Institute for Developmental Biology, Spemannstr. 35, 72076 Tübingen, Germany. kornelius.zeth@googlemail.com