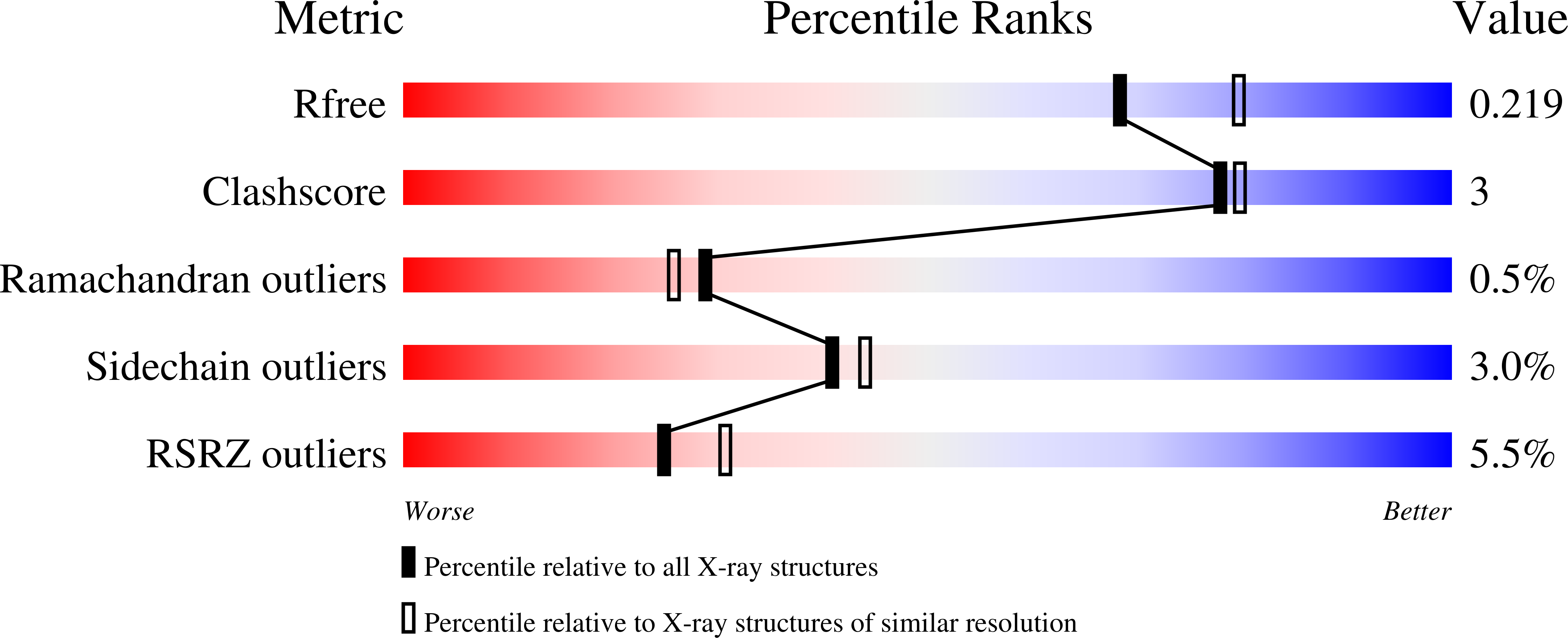
Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Barrault, M.B., Richet, N., Godard, C., Murciano, B., Le Tallec, B., Rousseau, E., Legrand, P., Charbonnier, J.B., Le Du, M.H., Guerois, R., Ochsenbein, F., Peyroche, A.(2012) Proc Natl Acad Sci U S A 109: E1001
- PubMed: 22460800
- DOI: https://doi.org/10.1073/pnas.1116538109
- Primary Citation of Related Structures:
4A3T, 4A3V - PubMed Abstract:
The 26S proteasome, a molecular machine responsible for regulated protein degradation, consists of a proteolytic core particle (20S CP) associated with 19S regulatory particles (19S RPs) subdivided into base and lid subcomplexes. The assembly of 19S RP base subcomplex is mediated by multiple dedicated chaperones. Among these, Hsm3 is important for normal growth and directly targets the carboxyl-terminal (C-terminal) domain of Rpt1 of the Rpt1-Rpt2-Rpn1 assembly intermediate. Here, we report crystal structures of the yeast Hsm3 chaperone free and bound to the C-terminal domain of Rpt1. Unexpectedly, the structure of the complex suggests that within the Hsm3-Rpt1-Rpt2 module, Hsm3 also contacts Rpt2. We show that in both yeast and mammals, Hsm3 actually directly binds the AAA domain of Rpt2. The Hsm3 C-terminal region involved in this interaction is required in vivo for base assembly, although it is dispensable for binding Rpt1. Although Rpt1 and Rpt2 exhibit weak affinity for each other, Hsm3 unexpectedly acts as an essential matchmaker for the Rpt1-Rpt2-Rpn1 assembly by bridging both Rpt1 and Rpt2. In addition, we provide structural and biochemical evidence on how Hsm3/S5b may regulate the 19S RP association to the 20S CP proteasome. Our data point out the diverse functions of assembly chaperones.
Organizational Affiliation:
Commissariat à l'Energie Atomique (CEA), Institut de Biologie et Technologies de Saclay (iBiTEC-S), Service de Biologie Intégrative et Génétique Moléculaire, Laboratoire du Métabolisme des Acides Nucléiques et Réponses aux Génotoxiques, F-91191 Gif-sur-Yvette, France.