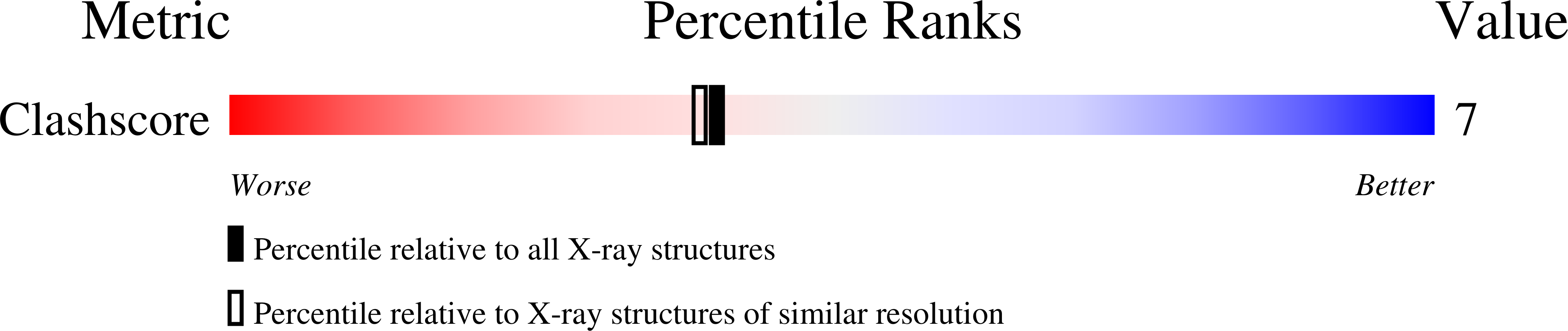
Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Selvakumar, P., Sharma, N., Tomar, P.P.S., Kumar, P., Sharma, A.K.(2014) Proteins 82: 830
- PubMed: 24265134
- DOI: https://doi.org/10.1002/prot.24461
- Primary Citation of Related Structures:
3ZC8, 3ZC9 - PubMed Abstract:
Murraya koenigii miraculin-like protein (MKMLP) gradually precipitates below pH 7.5. Here, we explore the basis for this aggregation by identifying the aggregation-prone regions via comparative analysis of crystal structures acquired at several pH values. The prediction of aggregation-prone regions showed the presence of four short peptides either in beta sheets or loops on surface of the protein. These peptides were distributed in two patches far apart on the surface. Comparison of crystal structures of MKMLP, determined at 2.2 Å resolution in pH 7.0 and 4.6 in the present study and determined at 2.9 Å in pH 8.0 in an earlier reported study, reveal subtle conformational differences resulting in gradual exposure of aggregation-prone regions. As the pH is lowered, there are alterations in ionic interactions within the protein interactions of the chain with water molecules and exposure of hydrophobic residues. The analysis of symmetry-related molecular interfaces involving one patch revealed shortening of nonpolar intermolecular contacts as the pH decreased. In particular, a decrease in the intermolecular distance between Trp103 of the aggregation-prone peptide WFITTG (103-108) unique to MLPs was observed. These results demonstrated that aggregation occurs due to the cumulative effect of the changes in interactions in two aggregation-prone defined regions.
Organizational Affiliation:
Department of Biotechnology, Indian Institute of Technology Roorkee, Roorkee, 247 667, India.