A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
Fukumoto, K., Onoda, A., Mizohata, E., Bocola, M., Inoue, T., Schwaneberg, U., Hayashi, T.(2014) ChemCatChem
Experimental Data Snapshot
(2014) ChemCatChem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UPF0678 fatty acid-binding protein-like protein At1g79260 | 174 | Arabidopsis thaliana | Mutation(s): 5 Gene Names: At1g79260, At1g79260.1, YUP8H12R.14 | 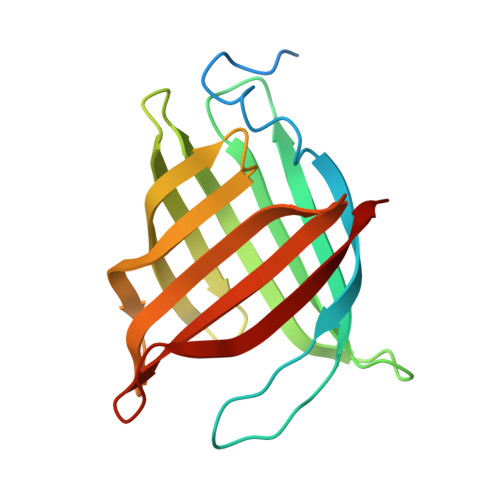 | |
UniProt | |||||
Find proteins for O64527 (Arabidopsis thaliana) Explore O64527 Go to UniProtKB: O64527 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O64527 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RMD Query on RMD | C [auth A], F [auth B] | [(1,2,5,6-eta)-cyclooctane-1,2,5,6-tetrayl]{(1,2,3,4,5-eta)-1-[2-(2,5-dioxopyrrolidin-1-yl)ethyl]cyclopentadienyl}rhodium C19 H24 N O2 Rh SZAWNSHHLUVDRC-UHFFFAOYSA-N |  | ||
| BA Query on BA | D [auth A], E [auth A], G [auth B], H [auth B] | BARIUM ION Ba XDFCIPNJCBUZJN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.888 | α = 90 |
| b = 67.888 | β = 90 |
| c = 129.895 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |