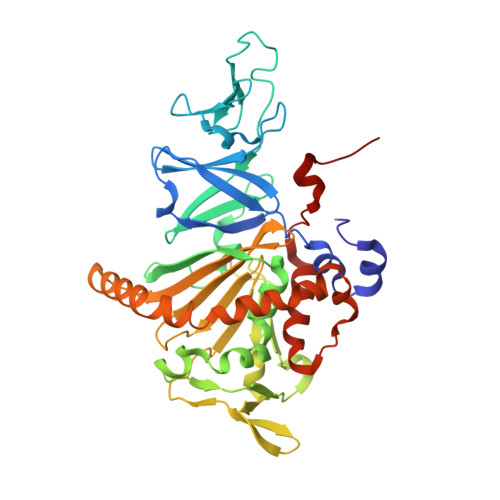
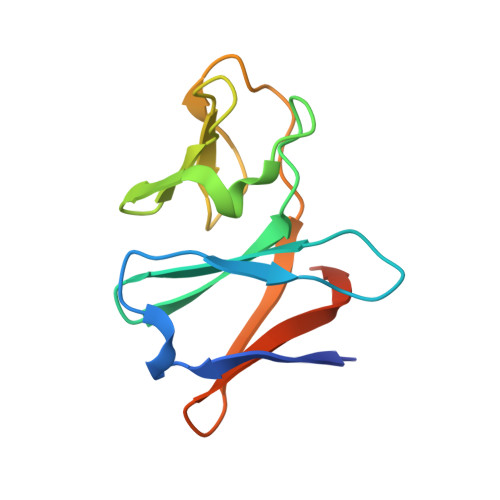
Structural insight into the substrate- and dioxygenbinding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9adioxygenase
Ashikawa, Y., Fujimoto, Z., Usami, Y., Inoue, K., Noguchi, H., Yamane, H., Nojiri, H.(2012) BMC Struct Biol 12: 15-15
- PubMed: 22727022
- DOI: https://doi.org/10.1186/1472-6807-12-15
- Primary Citation of Related Structures:
3VMG, 3VMH, 3VMI - PubMed Abstract:
Dihydroxylation of tandemly linked aromatic carbons in a cis-configuration, catalyzed by multicomponent oxygenase systems known as Rieske nonheme iron oxygenase systems (ROs), often constitute the initial step of aerobic degradation pathways for various aromatic compounds. Because such RO reactions inherently govern whether downstream degradation processes occur, novel oxygenation mechanisms involving oxygenase components of ROs (RO-Os) is of great interest. Despite substantial progress in structural and physicochemical analyses, no consensus exists on the chemical steps in the catalytic cycles of ROs. Thus, determining whether conformational changes at the active site of RO-O occur by substrate and/or oxygen binding is important. Carbazole 1,9a-dioxygenase (CARDO), a RO member consists of catalytic terminal oxygenase (CARDO-O), ferredoxin (CARDO-F), and ferredoxin reductase. We have succeeded in determining the crystal structures of oxidized CARDO-O, oxidized CARDO-F, and both oxidized and reduced forms of the CARDO-O: CARDO-F binary complex.
Organizational Affiliation:
Biotechnology Research Center, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.