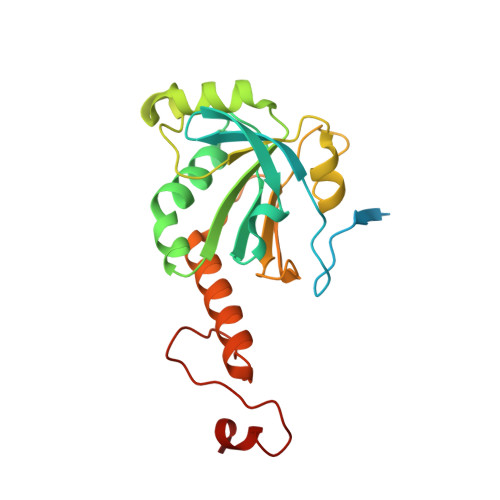
The N-terminal beta-sheet of peroxiredoxin Prx4 in the large yellow croaker Pseudosciaena crocea is critical for its peroxidase and anti-bacterial activities
Lian, F.M., Mu, Y.N., Teng, Y.B., Ao, J.Q., Jiang, Y.L., He, Y.X., Chen, Y.X., Zhou, C.Z., Chen, X.H.To be published.