Structure of Hibiscus latent Singapore virus by fiber diffraction: A non-conserved His122 contributes to coat protein stability
Tewary, S.K., Oda, T., Kendall, A., Bian, W., Stubbs, G., Wong, S.M., Swaminathan, K.(2010) J Mol Biol
- PubMed: 21195089
- DOI: https://doi.org/10.1016/j.jmb.2010.12.032
- Primary Citation of Related Structures:
3PDM - PubMed Abstract:
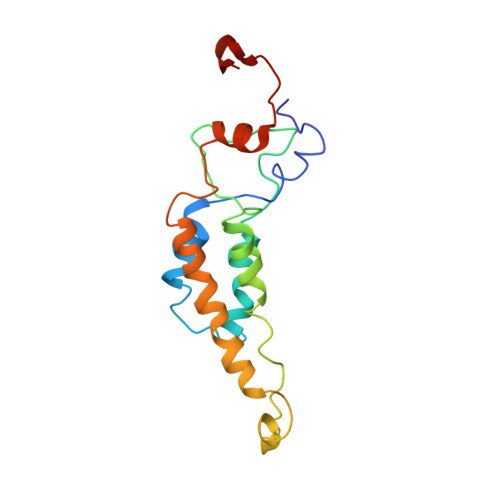
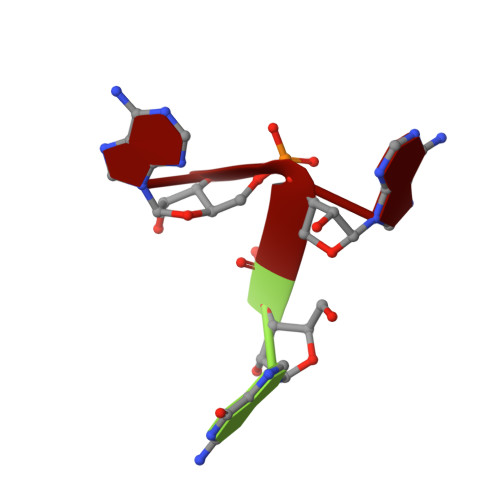
Hibiscus latent Singapore virus (HLSV) is a rigid rod-shaped plant virus and a new member of the Tobamovirus family. Unlike all other Tobamoviruses, the HLSV genome contains a unique poly(A) tract in its 3' untranslated region. The virion is composed of a monomeric coat protein (CP) unit of 18 kDa, arranged as a right-handed helix around the virus axis. We have determined the structure of HLSV at 3.5 Å by X-ray fiber diffraction and refined it to an R-factor of 0.096. While the overall structure of the HLSV CP resembles that of other Tobamoviruses, there are a few unique differences. There is a kink in the LR helix due to the presence of His122. Also, the adjacent Lys123 may further destabilize the helix by positive charge repulsion, making the kink more pronounced. The His122-Asp88 salt bridge provides significant stability to the loop adjacent to the RR helix. Carboxyl-carboxylate interactions that drive viral disassembly are also different in HLSV. The nucleotide recognition mechanisms for virus assembly between HLSV and ribgrass mosaic virus are similar, but different between tobacco mosaic virus and cucumber green mottle mosaic virus.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.