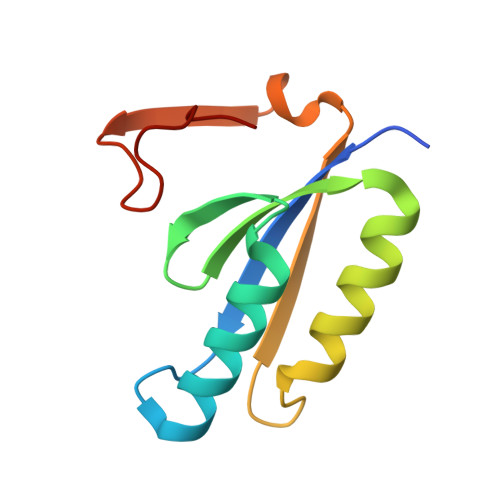
Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris.
Samai, P., Smith, P., Shuman, S.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1552-1556
- PubMed: 21139194
- DOI: https://doi.org/10.1107/S1744309110039801
- Primary Citation of Related Structures:
3OQ2 - PubMed Abstract:
CRISPRs (clustered regularly interspaced short palindromic repeats) provide bacteria and archaea with RNA-guided acquired immunity to invasive DNAs. CRISPR-associated (Cas) proteins carry out the immune effector functions. Cas2 is a universal component of the CRISPR system. Here, a 1.35 Å resolution crystal structure of Cas2 from the bacterium Desulfovibrio vulgaris (DvuCas2) is reported. DvuCas2 is a homodimer, with each protomer consisting of an N-terminal βαββαβ ferredoxin fold (amino acids 1-78) to which is appended a C-terminal segment (amino acids 79-102) that includes a short 3(10)-helix and a fifth β-strand. The β5 strands align with the β4 strands of the opposite protomers, resulting in two five-stranded antiparallel β-sheets that form a sandwich at the dimer interface. The DvuCas2 dimer is stabilized by a distinctive network of hydrophilic cross-protomer side-chain interactions.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute for Cancer Research, USA.