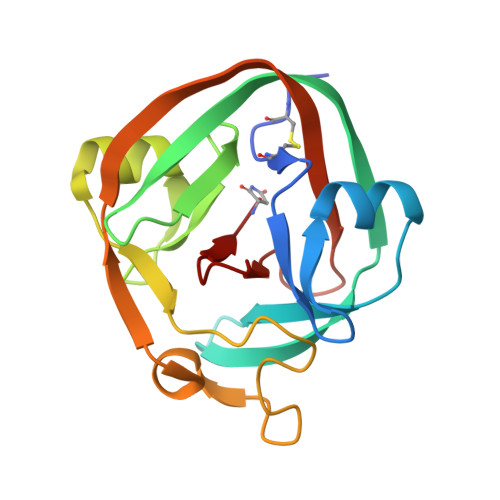
Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications.
Callahan, B.P., Topilina, N.I., Stanger, M.J., Van Roey, P., Belfort, M.(2011) Nat Struct Mol Biol 18: 630-633
- PubMed: 21460844
- DOI: https://doi.org/10.1038/nsmb.2041
- Primary Citation of Related Structures:
3NZM - PubMed Abstract:
Here we describe self-splicing proteins, called inteins, that function as redox-responsive switches in bacteria. Redox regulation was achieved by engineering a disulfide bond between the intein's catalytic cysteine and a cysteine in the flanking 'extein' sequence. This interaction was validated by an X-ray structure, which includes a transient splice junction. A natural analog of the designed system was identified in Pyrococcus abyssi, suggesting an unprecedented form of adaptive, post-translational regulation.
Organizational Affiliation:
Wadsworth Center, New York State Department of Health, Albany, New York, USA.