Crystal Structure of d(CACGCG).d(CGCGTG) with 10mM MnCl2
Venkadesh, S., Mandal, P.K., Gautham, N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*CP*AP*CP*GP*CP*G)-3' | 6 | synthetic construct | 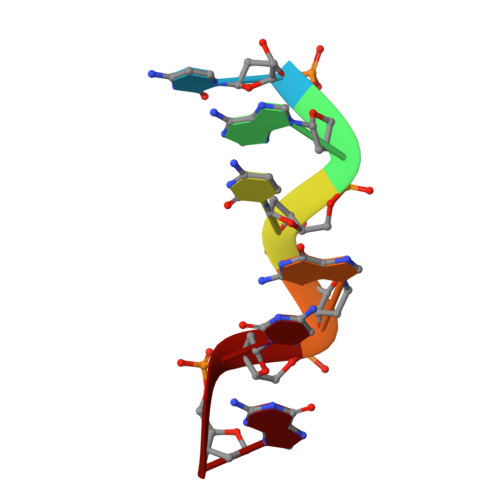 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-D(*CP*GP*CP*GP*TP*G)-3' | 6 | synthetic construct |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MN Query on MN | E [auth A], F [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 24.612 | α = 90 |
| b = 31.257 | β = 103.64 |
| c = 31.962 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| AUTOMAR | data reduction |
| SCALEPACK | data scaling |