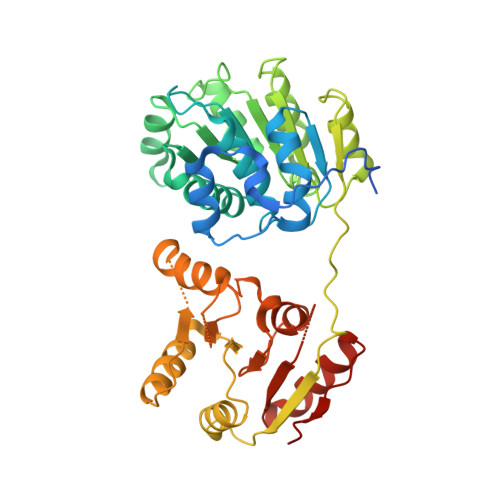
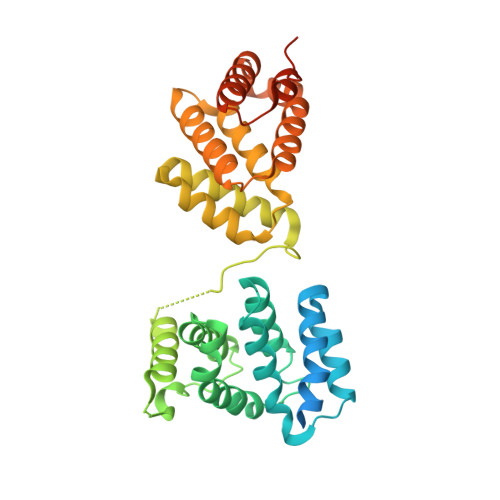
Structural basis for translational inhibition by the tumour suppressor Pdcd4
Loh, P.G., Yang, H.S., Walsh, M.A., Wang, Q., Wang, X., Cheng, Z., Liu, D., Song, H.(2009) EMBO J 28: 274-285
- PubMed: 19153607
- DOI: https://doi.org/10.1038/emboj.2008.278
- Primary Citation of Related Structures:
3EIJ, 3EIQ - PubMed Abstract:
Pdcd4 is a tumour suppressor protein. It inhibits translation through interaction with translation initiator eIF4A, resulting in the suppression of neoplastic transformation and tumour invasion. Here, we present the crystal structures of an N-terminal-truncated Pdcd4 in free form and in complex with eIF4A. Upon binding to eIF4A, Pdcd4 undergoes a marked conformational change to form a heterotrimeric complex with eIF4A, with one Pdcd4 binding to two eIF4A molecules in two different modes. The binding of Pdcd4 to eIF4A is required to inhibit the enzymatic activity of eIF4A, translation initiation, and AP-1-dependent transcription. Both MA3 domains are required to efficiently compete with the C-terminal domain of eIF4G (eIF4Gc) for binding to eIF4A whereas a single MA3 is sufficient to inhibit translation. Our structural and mutational analyses reveal that Pdcd4 inhibits translation initiation by trapping eIF4A in an inactive conformation, and blocking its incorporation into the eIF4F complex.
Organizational Affiliation:
Institute of Molecular and Cell Biology, Proteos, Singapore.