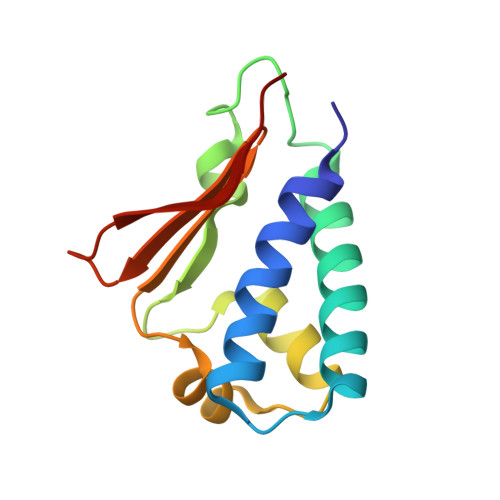
Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Yang, X., Morita, M., Wang, H., Suzuki, T., Yang, W., Luo, Y., Zhao, C., Yu, Y., Bartlam, M., Yamamoto, T., Rao, Z.(2008) Nucleic Acids Res 36: 6872-6881
- PubMed: 18974182
- DOI: https://doi.org/10.1093/nar/gkn825
- Primary Citation of Related Structures:
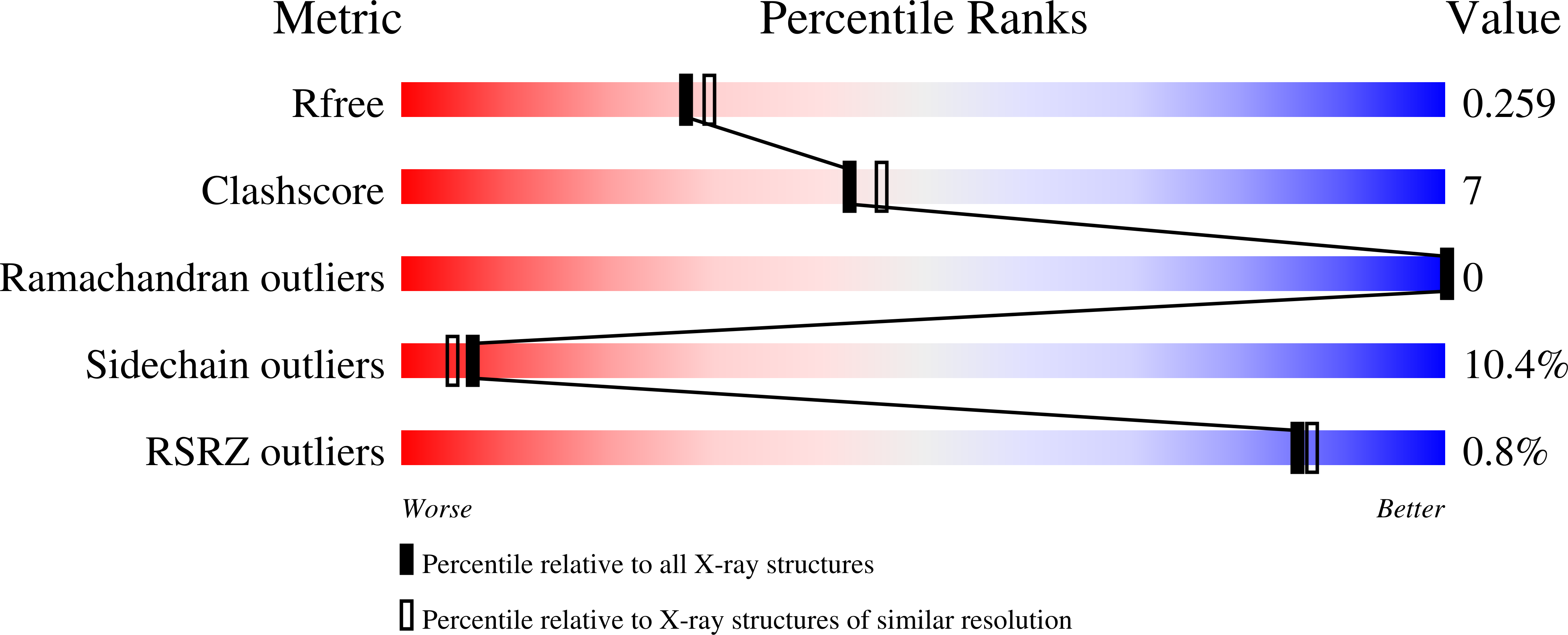
3DJN, 3DJU - PubMed Abstract:
BTG2 is the prototypical member of the TOB family and is known to be involved in cell growth, differentiation and DNA repair. As a transcriptional co-regulator, BTG2 interacts with CCR4-associated factor 1 (CAF1) and POP2 (CALIF), which are key components of the general CCR4/NOT multi-subunit transcription complex, and which are reported to play distinct roles as nucleases involved in mRNA deadenylation. Here we report the crystal structures of human BTG2 and mouse TIS21 to 2.3 A and 2.2 A resolution, respectively. The structures reveal the putative CAF1 binding site. CAF1 deadenylase assays were performed with wild-type BTG2 and mutants that disrupt the interaction with CAF1. The results reveal the suppressive role of BTG2 in the regulation of CAF1 deadenylase activity. Our study provides insights into the formation of the BTG2-CAF1 complex and the potential role of BTG2 in the regulation of CAF1.
Organizational Affiliation:
Laboratory of Structural Biology, Tsinghua University, Beijing, China.