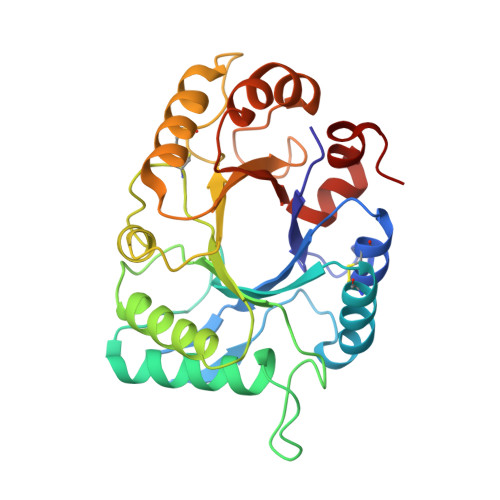
Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance
Kumar, S., Singh, N., Sinha, M., Singh, S.B., Bhushan, A., Kaur, P., Srinivasan, A., Sharma, S., Singh, T.P.To be published.