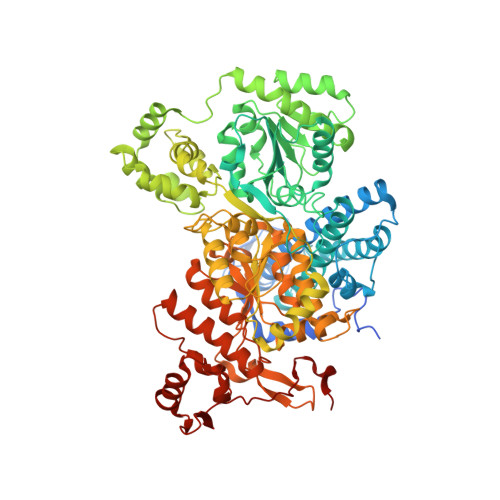
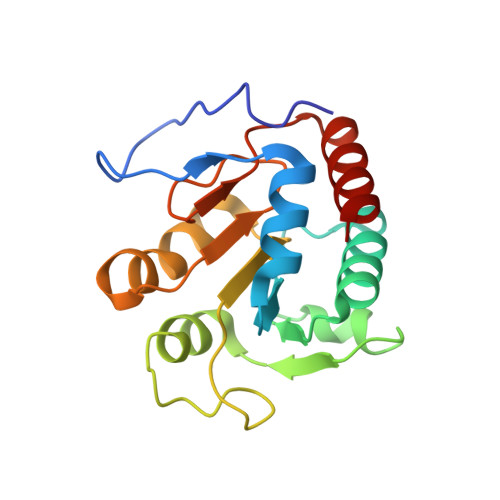
Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Gong, W., Hao, B., Wei, Z., Ferguson Jr., D.J., Tallant, T., Krzycki, J.A., Chan, M.K.(2008) Proc Natl Acad Sci U S A 105: 9558-9563
- PubMed: 18621675
- DOI: https://doi.org/10.1073/pnas.0800415105
- Primary Citation of Related Structures:
3CF4 - PubMed Abstract:
Ni-dependent carbon monoxide dehydrogenases (Ni-CODHs) are a diverse family of enzymes that catalyze reversible CO:CO(2) oxidoreductase activity in acetogens, methanogens, and some CO-using bacteria. Crystallography of Ni-CODHs from CO-using bacteria and acetogens has revealed the overall fold of the Ni-CODH core and has suggested structures for the C cluster that mediates CO:CO(2) interconversion. Despite these advances, the mechanism of CO oxidation has remained elusive. Herein, we report the structure of a distinct class of Ni-CODH from methanogenic archaea: the alpha(2)epsilon(2) component from the alpha(8)beta(8)gamma(8)delta(8)epsilon(8) CODH/acetyl-CoA decarbonylase/synthase complex, an enzyme responsible for the majority of biogenic methane production on Earth. The structure of this Ni-CODH component provides support for a hitherto unobserved state in which both CO and H(2)O/OH(-) bind to the Ni and the exogenous FCII iron of the C cluster, respectively, and offers insight into the structures and functional roles of the epsilon-subunit and FeS domain not present in nonmethanogenic Ni-CODHs.
Organizational Affiliation:
Departments of Biochemistry, Chemistry and Microbiology, and Ohio State Biochemistry Program, Ohio State University, 484 West 12th Avenue, Columbus, OH 43210, USA.