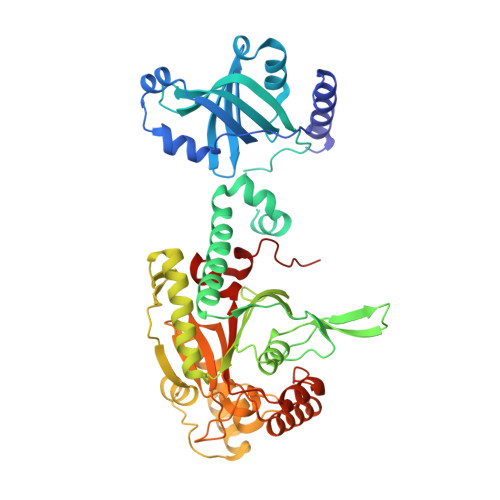
Crystal Structure of Lysyl-tRNA Synthetase from Bacillus stearothermophilus in Complex with Diadenosine Tetraphosphate (AP4A): Insights into AP4A Synthesis Mechanisms and Implication for Recognition of Discriminator Base of tRNA^Lys
Sakurama, H., Takita, T., Mikami, B., Itoh, T., Yasukawa, K., Inouye, K.To be published.