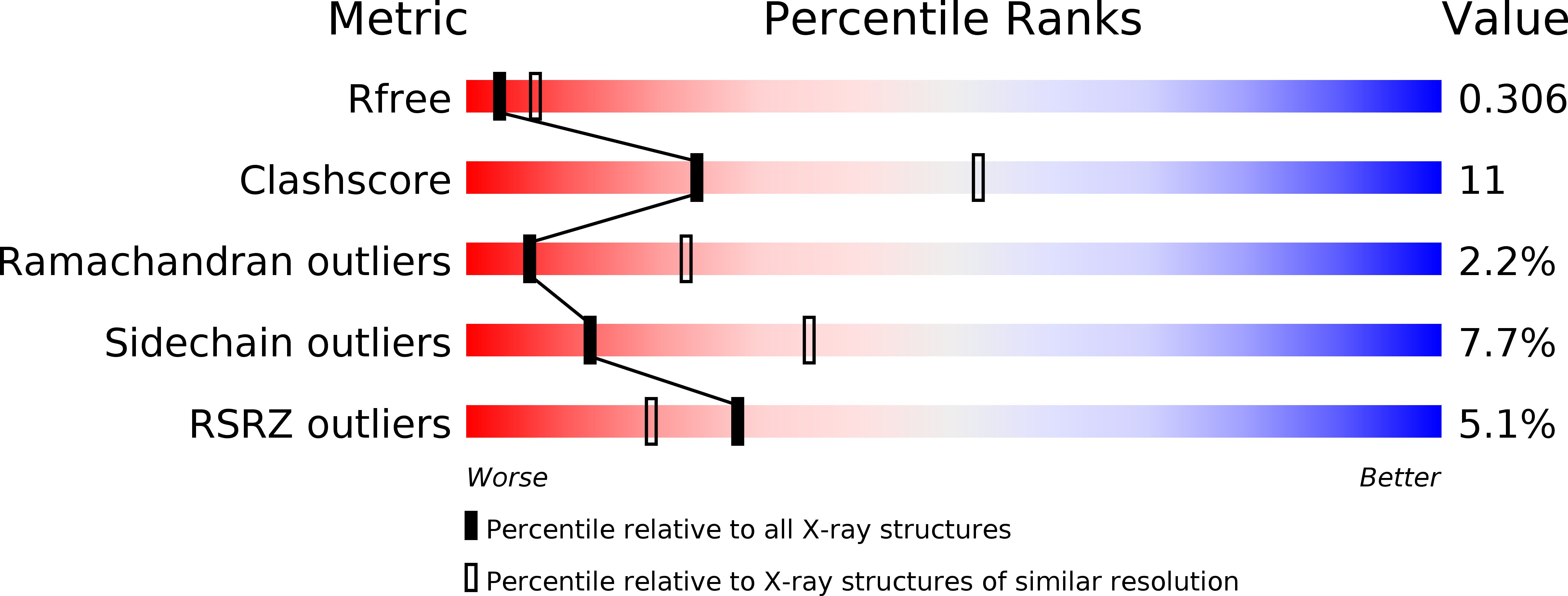
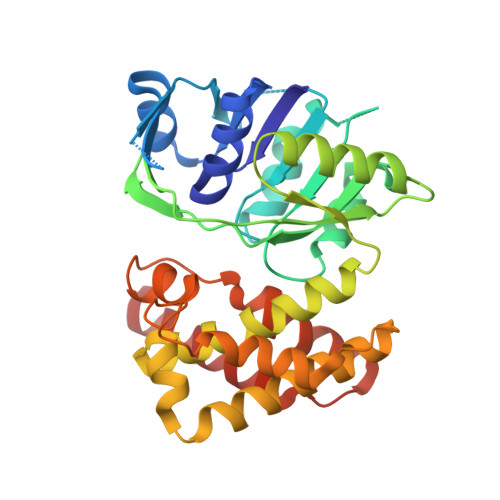
The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Miyanaga, A., Fujisawa, S., Furukawa, N., Arai, K., Nakajima, M., Taguchi, H.(2013) Biochem Biophys Res Commun 439: 109-114
- PubMed: 23954635
- DOI: https://doi.org/10.1016/j.bbrc.2013.08.019
- Primary Citation of Related Structures:
3WFI, 3WFJ - PubMed Abstract:
D-Mandelate dehydrogenases (D-ManDHs), belonging to a new d-2-hydroxyacid dehydrogenase family, catalyze the conversion between benzoylformate and d-mandelate using NAD as a coenzyme. We determined the first D-ManDH structure, that of ManDH2 from Enterococcus faecalis IAM10071. The overall structure showed ManDH2 has a similar fold to 2-ketopantoate reductase (KPR), which catalyzes the conversion of 2-ketopantoate to d-pantoate using NADP as a coenzyme. They share conserved catalytic residues, indicating ManDH2 has the same reaction mechanism as KPR. However, ManDH2 exhibits significant structural variations in the coenzyme and substrate binding sites compared to KPR. These structural observations could explain their different coenzyme and substrate specificities.
Organizational Affiliation:
Department of Applied Biological Science, Faculty of Science and Technology, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan. Electronic address: miyanaga.a.aa@m.titech.ac.jp.