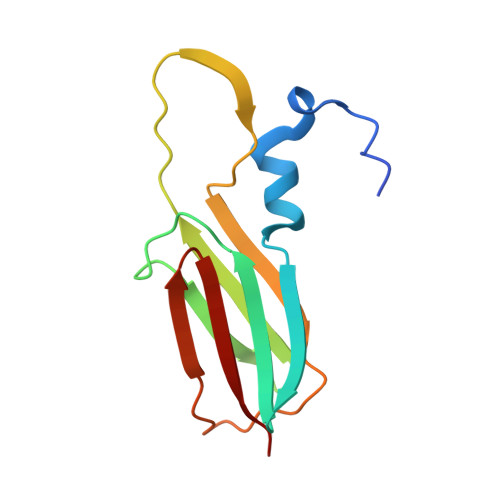
Structural studies on the oligomeric transition of a small heat shock protein, StHsp14.0
Hanazono, Y., Takeda, K., Yohda, M., Miki, K.(2012) J Mol Biol 422: 100-108
- PubMed: 22613762
- DOI: https://doi.org/10.1016/j.jmb.2012.05.017
- Primary Citation of Related Structures:
3VQK, 3VQL, 3VQM - PubMed Abstract:
The small heat shock proteins (sHsps), which are widely found in all domains of life, bind and stabilize denatured proteins to prevent aggregation. The sHsps exist as large oligomers that are composed of 9-40 subunits and control their chaperone activity by the transition of the oligomeric state. Though the oligomeric transition is important for the biological function of most sHsps, atomic details have not been elucidated. Here, we report crystal structures in both the 24-meric and dimeric states for an sHsp, StHsp14.0 from Sulfolobus tokodaii, in order to reveal changes upon the oligomeric transition. The results indicate that StHsp14.0 forms a spherical 24-mer with a diameter of 115 Å. The diameter is defined by the inter-monomer angle in the dimer. The dimer structure in the dimeric state shows only small differences from that in the 24-meric state. Some significant differences are exclusively observed at the binding site for the C-terminus. Although a dimer has four interactive sites with neighboring dimers, the weakness of the respective interactions is indicated from the size-exclusion chromatography. The small structural changes imply an activation mechanism mediated by multiple weak interactions.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan.