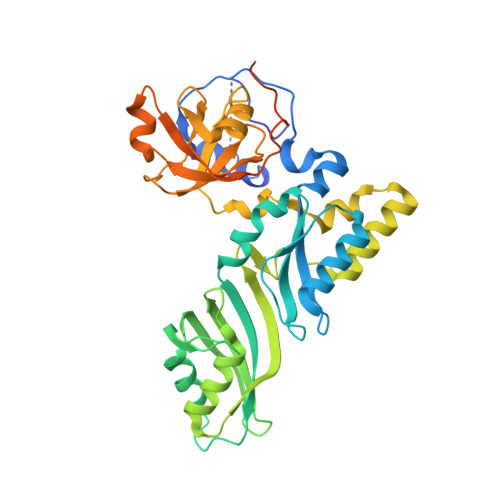
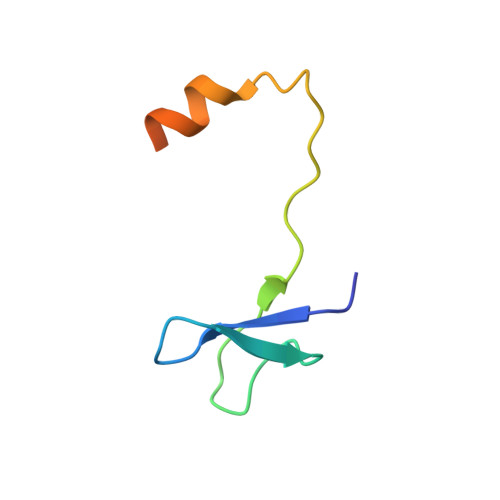
Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase
Li, S., Duan, J., Li, D., Yang, B., Dong, M., Ye, K.(2011) Genes Dev 25: 2409-2421
- PubMed: 22085967
- DOI: https://doi.org/10.1101/gad.175299.111
- Primary Citation of Related Structures:
3U28 - PubMed Abstract:
Box H/ACA ribonucleoprotein particles (RNPs) mediate pseudouridine synthesis, ribosome formation, and telomere maintenance. The structure of eukaryotic H/ACA RNPs remains poorly understood. We reconstituted functional Saccharomyces cerevisiae H/ACA RNPs with recombinant proteins Cbf5, Nop10, Gar1, and Nhp2 and a two-hairpin H/ACA RNA; determined the crystal structure of a Cbf5, Nop10, and Gar1 ternary complex at 1.9 Å resolution; and analyzed the structure-function relationship of the yeast complex. Although eukaryotic H/ACA RNAs have a conserved two-hairpin structure, isolated single-hairpin RNAs are also active in guiding pseudouridylation. Nhp2, unlike its archaeal counterpart, is largely dispensable for the activity, reflecting a functional adaptation of eukaryotic H/ACA RNPs to the variable RNA structure that Nhp2 binds. The N-terminal extension of Cbf5, a hot spot for dyskeratosis congenita mutation, forms an extra structural layer on the PUA domain. Gar1 is distinguished from the assembly factor Naf1 by containing a C-terminal extension that controls substrate turnover and the Gar1-Naf1 exchange during H/ACA RNP maturation. Our results reveal significant novel features of eukaryotic H/ACA RNPs.
Organizational Affiliation:
National Institute of Biological Sciences, Beijing, China.