Structure and atypical hydrolysis mechanism of the Nudix hydrolase Orf153 (YmfB) from Escherichia coli
Hong, M.K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Biological assembly 1 assigned by authors.
Biological assembly 2 assigned by authors.
Biological assembly 3 assigned by authors.
Biological assembly 4 assigned by authors.
Biological assembly 5 assigned by authors.
Biological assembly 6 assigned by authors.
Biological assembly 7 assigned by authors.
Biological assembly 8 assigned by authors.
Biological assembly 9 assigned by authors.
Biological assembly 10 assigned by authors.
Biological assembly 11 assigned by authors.
Biological assembly 12 assigned by authors.
Biological assembly 13 generated by PISA (software)
Biological assembly 14 generated by PISA (software)
Biological assembly 15 generated by PISA (software)
Biological assembly 16 generated by PISA (software)
Biological assembly 17 generated by PISA (software)
Biological assembly 18 generated by PISA (software)
Macromolecule Content
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphatase nudJ | 153 | Escherichia coli APEC O1 | Mutation(s): 0 Gene Names: nudJ, Ecok1_10240, APECO1_216 EC: 3.6.1 | 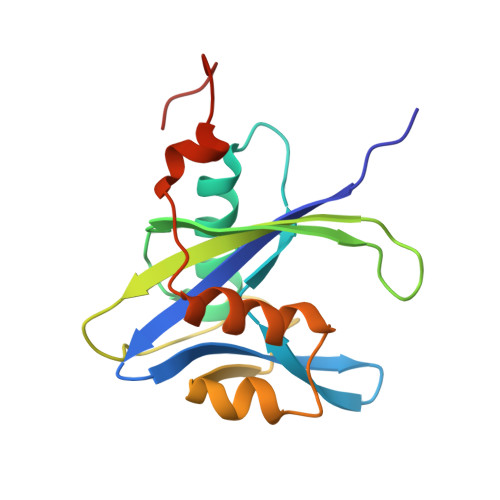 | |
UniProt | |||||
Find proteins for A1AA28 (Escherichia coli O1:K1 / APEC) Explore A1AA28 Go to UniProtKB: A1AA28 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1AA28 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 111.126 | α = 90 |
| b = 111.126 | β = 90 |
| c = 247.306 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASES | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
RCSB PDB (citation) is hosted by
RCSB PDB Core Operations are funded by the National Science Foundation (DBI-1832184), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM133198.


