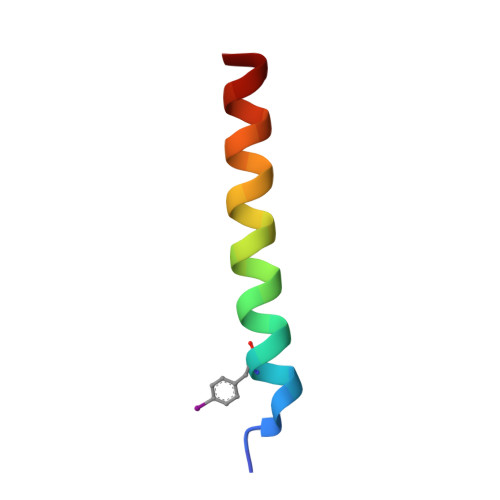
Cryo-transmission electron microscopy structure of a gigadalton peptide fiber of de novo design
Sharp, T.H., Bruning, M., Mantell, J., Sessions, R.B., Thomson, A.R., Zaccai, N.R., Brady, R.L., Verkade, P., Woolfson, D.N.(2012) Proc Natl Acad Sci U S A 109: 13266-13271
- PubMed: 22847414
- DOI: https://doi.org/10.1073/pnas.1118622109
- Primary Citation of Related Structures:
3RA3 - PubMed Abstract:
Nature presents various protein fibers that bridge the nanometer to micrometer regimes. These structures provide inspiration for the de novo design of biomimetic assemblies, both to address difficulties in studying and understanding natural systems, and to provide routes to new biomaterials with potential applications in nanotechnology and medicine. We have designed a self-assembling fiber system, the SAFs, in which two small α-helical peptides are programmed to form a dimeric coiled coil and assemble in a controlled manner. The resulting fibers are tens of nm wide and tens of μm long, and, therefore, comprise millions of peptides to give gigadalton supramolecular structures. Here, we describe the structure of the SAFs determined to approximately 8 Å resolution using cryotransmission electron microscopy. Individual micrographs show clear ultrastructure that allowed direct interpretation of the packing of individual α-helices within the fibers, and the construction of a 3D electron density map. Furthermore, a model was derived using the cryotransmission electron microscopy data and side chains taken from a 2.3 Å X-ray crystal structure of a peptide building block incapable of forming fibers. This was validated using single-particle analysis techniques, and was stable in prolonged molecular-dynamics simulation, confirming its structural viability. The level of self-assembly and self-organization in the SAFs is unprecedented for a designed peptide-based material, particularly for a system of considerably reduced complexity compared with natural proteins. This structural insight is a unique high-resolution description of how α-helical fibrils pack into larger protein fibers, and provides a basis for the design and engineering of future biomaterials.
Organizational Affiliation:
School of Chemistry, University of Bristol, Cantock's Close, Bristol BS8 1TS, United Kingdom.