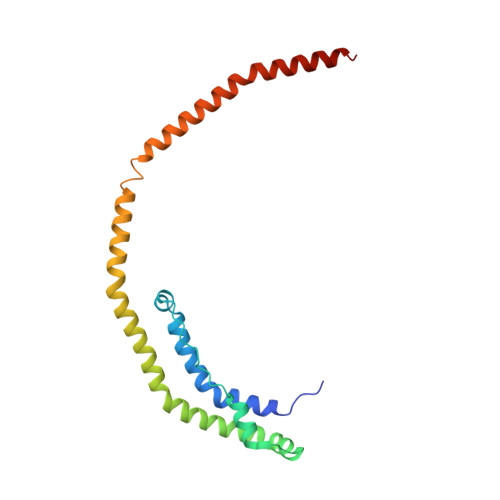
Crystal Structure of C-terminal Truncated Apolipoprotein A-I Reveals the Assembly of High Density Lipoprotein (HDL) by Dimerization.
Mei, X., Atkinson, D.(2011) J Biol Chem 286: 38570-38582
- PubMed: 21914797
- DOI: https://doi.org/10.1074/jbc.M111.260422
- Primary Citation of Related Structures:
3R2P - PubMed Abstract:
Apolipoprotein A-I (apoA-I) plays important structural and functional roles in plasma high density lipoprotein (HDL) that is responsible for reverse cholesterol transport. However, a molecular understanding of HDL assembly and function remains enigmatic. The 2.2-Å crystal structure of Δ(185-243)apoA-I reported here shows that it forms a half-circle dimer. The backbone of the dimer consists of two elongated antiparallel proline-kinked helices (five AB tandem repeats). The N-terminal domain of each molecule forms a four-helix bundle with the helical C-terminal region of the symmetry-related partner. The central region forms a flexible domain with two antiparallel helices connecting the bundles at each end. The two-domain dimer structure based on helical repeats suggests the role of apoA-I in the formation of discoidal HDL particles. Furthermore, the structure suggests the possible interaction with lecithin-cholesterol acyltransferase and may shed light on the molecular details of the effect of the Milano, Paris, and Fin mutations.
Organizational Affiliation:
Department of Physiology and Biophysics, Boston University School of Medicine, Boston, Massachusetts 02118.