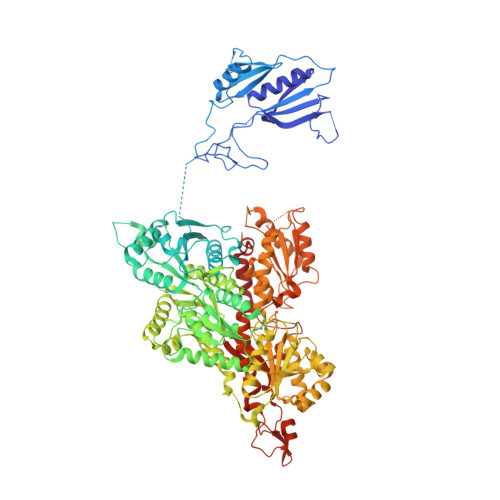
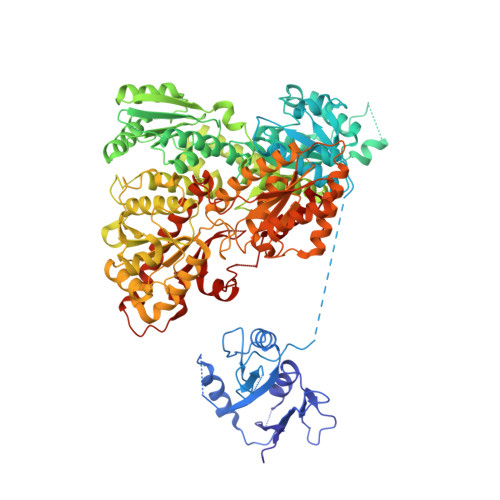
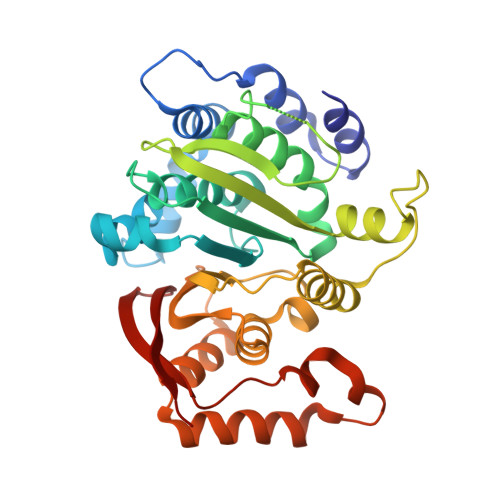
Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Strater, N., Marek, S., Kuettner, E.B., Kloos, M., Keim, A., Bruser, A., Kirchberger, J., Schoneberg, T.(2011) FASEB J 25: 89-98
- PubMed: 20833871
- DOI: https://doi.org/10.1096/fj.10-163865
- Primary Citation of Related Structures:
3OPY - PubMed Abstract:
Eukaryotic ATP-dependent 6-phosphofructokinases (Pfks) differ from their bacterial counterparts in a much more complex structural organization and allosteric regulation. Pichia pastoris Pfk (PpPfk) is, with ∼ 1 MDa, the most complex and probably largest eukaryotic Pfk. We have determined the crystal structure of full-length PpPfk to 3.05 Å resolution in the T state. PpPfk forms a (αβγ)(4) dodecamer of D(2) symmetry with dimensions of 161 × 157 × 233 Å mainly via interactions of the α chains. The N-terminal domains of the α and β chains have folds that are distantly related to glyoxalase I, but the active sites are no longer functional. Interestingly, these domains located at the 2 distal ends of this protein along the long 2-fold axis form a (αβ)(2) dimer as does the core Pfk domains; however, the domains are swapped across the tetramerization interface. In PpPfk, the unique γ subunit participates in oligomerization of the αβ chains. This modulator protein was acquired from an ancient S-adenosylmethionine-dependent methyltransferase. The identification of novel ATP binding sites, which do not correspond to the bacterial catalytic or effector binding sites, point to marked structural and functional differences between bacterial and eukaryotic Pfks.
Organizational Affiliation:
Institute of Bioanalytical Chemistry, Center for Biotechnology and Biomedicine, University of Leipzig, Deutscher Platz 5, 04103 Leipzig, Germany. strater@bbz.uni-leipzig.de