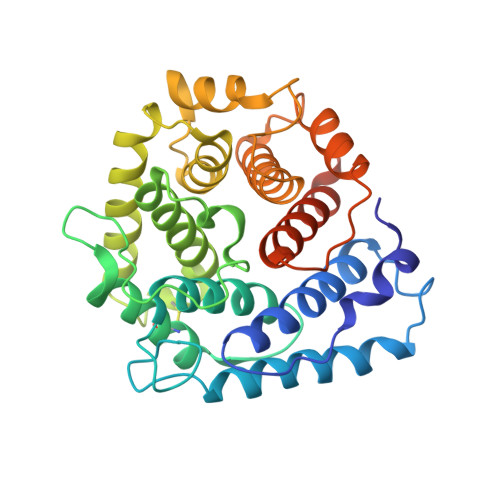
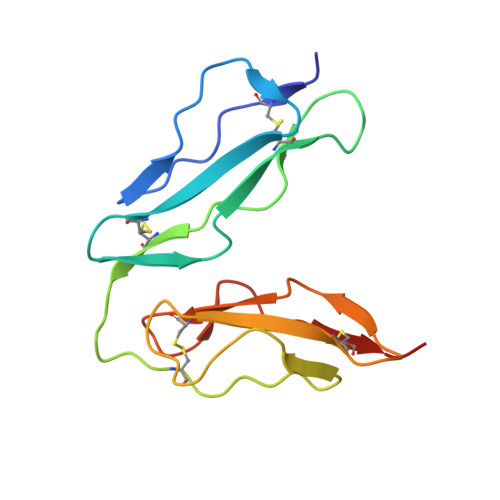
A crystal structure of the complex between human complement receptor 2 and its ligand C3d.
van den Elsen, J.M., Isenman, D.E.(2011) Science 332: 608-611
- PubMed: 21527715
- DOI: https://doi.org/10.1126/science.1201954
- Primary Citation of Related Structures:
3OED - PubMed Abstract:
The interaction of complement receptor 2 (CR2)--which is present on B cells and follicular dendritic cells--with its antigen-bound ligand C3d results in an enhanced antibody response, thus providing an important link between the innate and adaptive immune systems. Although a cocrystal structure of a complex between C3d and the ligand-binding domains of CR2 has been published, several aspects of this structure, including the position in C3d of the binding interface, remained controversial because of disagreement with biochemical data. We now report a cocrystal structure of a CR2(SCR1-2):C3d complex at 3.2 angstrom resolution in which the interaction interfaces differ markedly from the previously published structure and are consistent with the biochemical data. It is likely that, in the previous structure, the interaction was influenced by the presence of zinc acetate additive in the crystallization buffer, leading to a nonphysiological complex. Detailed knowledge of the binding interface now at hand gives the potential to exploit the interaction in vaccine design or in therapeutics directed against autoreactive B cells.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Bath BA2 7AY, UK. bssjmhve@bath.ac.uk