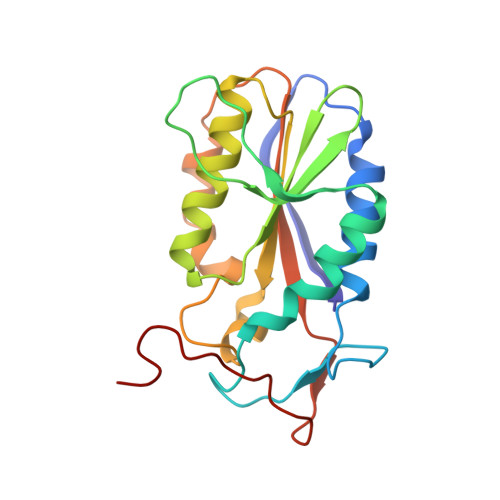
Asymmetric DNA recognition by the OkrAI endonuclease, an isoschizomer of BamHI.
Vanamee, E.S., Viadiu, H., Chan, S.H., Ummat, A., Hartline, A.M., Xu, S.Y., Aggarwal, A.K.(2011) Nucleic Acids Res 39: 712-719
- PubMed: 20833632
- DOI: https://doi.org/10.1093/nar/gkq779
- Primary Citation of Related Structures:
3ODH - PubMed Abstract:
Restriction enzymes share little or no sequence homology with the exception of isoschizomers, or enzymes that recognize and cleave the same DNA sequence. We present here the structure of a BamHI isoschizomer, OkrAI, bound to the same DNA sequence (TATGGATCCATA) as that cocrystallized with BamHI. We show that OkrAI is a more minimal version of BamHI, lacking not only the N- and C-terminal helices but also an internal 3(10) helix and containing β-strands that are shorter than those in BamHI. Despite these structural differences, OkrAI recognizes the DNA in a remarkably similar manner to BamHI, including asymmetric contacts via C-terminal 'arms' that appear to 'compete' for the minor groove. However, the arms are shorter than in BamHI. We observe similar DNA-binding affinities between OkrAI and BamHI but OkrAI has higher star activity (at 37°C) compared to BamHI. Together, the OkrAI and BamHI structures offer a rare opportunity to compare two restriction enzymes that work on exactly the same DNA substrate.
Organizational Affiliation:
Department of Structural and Chemical Biology, Mount Sinai School of Medicine, New York, NY 10029, USA.