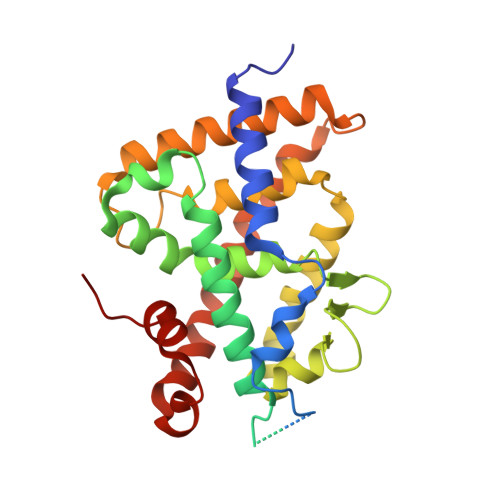
Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097.
Huet, T., Maehr, H., Lee, H.J., Uskokovic, M.R., Suh, N., Moras, D., Rochel, N.(2011) Medchemcomm 2: 424-429
- PubMed: 22180837
- DOI: https://doi.org/10.1039/C1MD00059D
- Primary Citation of Related Structures:
3O1D, 3O1E - PubMed Abstract:
Derivatives of vitamin D(3) containing a second side-chain emanating at C-20 are known as gemini and act as vitamin D receptor agonists. Recently, two of these, namely Gemini-0072 and the epimeric Gemini-0097, were selected for further studies in view of their high biological activities and lack of hypercalcemic effects. We now show that the two analogs recruit coactivator SRC-1 better than the parental gemini and act as VDR superagonists. The crystal structures of complexes of zVDR with Gemini-0072 and Gemini-0097 indicate that these ligands induce an extra cavity within the ligand-binding pocket similar to gemini and that their superagonistic activity is due to an increased stabilization of helix H12.
Organizational Affiliation:
Département de Biologie et de Génomique Structurales, IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Centre National de la Recherche Scientifique, Institut National de la Santé de la Recherche Méedicale, Université de Strasbourg, 1 rue Laurent Fries, 67404 Illkirch, France.