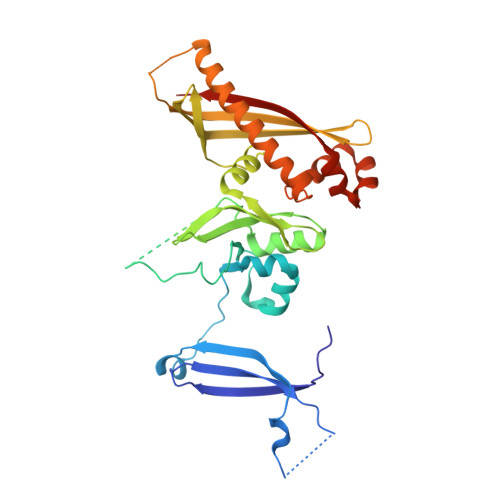
Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
Li, Y., Setlow, B., Setlow, P., Hao, B.(2010) J Mol Biol 402: 8-16
- PubMed: 20654628
- DOI: https://doi.org/10.1016/j.jmb.2010.07.018
- Primary Citation of Related Structures:
3N54 - PubMed Abstract:
The nutrient germinant receptors (nGRs) of spores of Bacillus species are clusters of three proteins that play a critical role in triggering the germination of dormant spores in response to specific nutrient molecules. Here, we report the crystal structure of the C protein of the GerB germinant receptor, so-called GerBC, of Bacillus subtilis spores at 2.3 A resolution. The GerBC protein adopts a previously uncharacterized type of protein fold consisting of three distinct domains, each of which is centered by a beta sheet surrounded by multiple alpha helices. Secondary-structure prediction and structure-based sequence alignment suggest that the GerBC structure represents the prototype for C subunits of nGRs from spores of all Bacillales and Clostridiales species and defines two highly conserved structural regions in this family of proteins. GerBC forms an interlocked dimer in the crystalline state but is predominantly monomeric in solution, pointing to the possibility that GerBC oligomerizes as a result of either high local protein concentrations or interaction with other nGR proteins in spores. Our findings provide the first structural view of the nGR subunits and a molecular framework for understanding the architecture, conservation, and function of nGRs.
Organizational Affiliation:
Department of Molecular,Microbial and Structural Biology, University of Connecticut Health Center, Farmington, CT 06030-3305, USA.