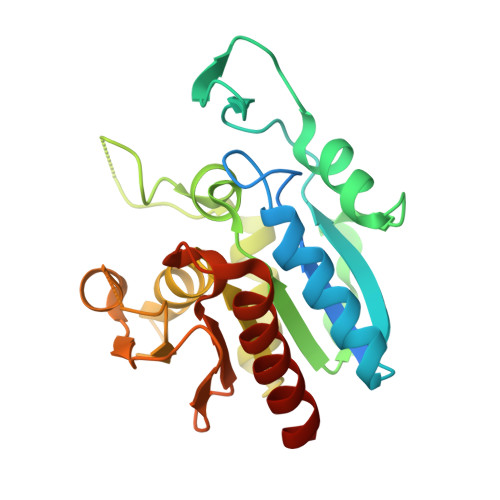
Crystal structure of isoprenoid biosynthesis protein with amidotransferase-like domain from Ehrlichia Chaffeensis at 1.90A resolution
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J., Sankaran, B., Edwards, T.E., Staker, B.To be published.