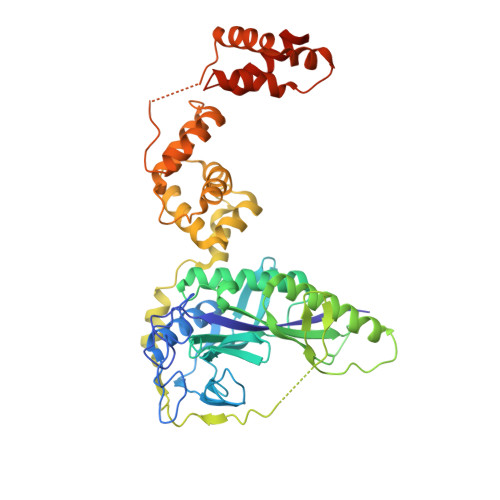
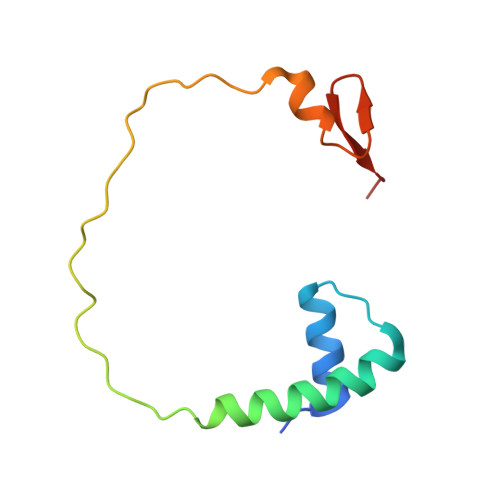
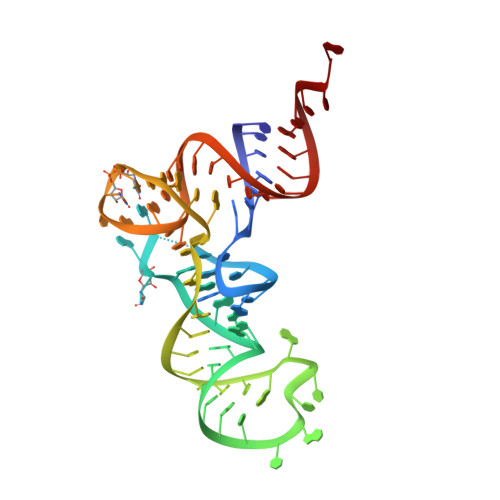
Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
Blaise, M., Bailly, M., Frechin, M., Behrens, M.A., Fischer, F., Oliveira, C.L., Becker, H.D., Pedersen, J.S., Thirup, S., Kern, D.(2010) EMBO J 29: 3118-3129
- PubMed: 20717102
- DOI: https://doi.org/10.1038/emboj.2010.192
- Primary Citation of Related Structures:
3KFU - PubMed Abstract:
Four out of the 22 aminoacyl-tRNAs (aa-tRNAs) are systematically or alternatively synthesized by an indirect, two-step route requiring an initial mischarging of the tRNA followed by tRNA-dependent conversion of the non-cognate amino acid. During tRNA-dependent asparagine formation, tRNA(Asn) promotes assembly of a ribonucleoprotein particle called transamidosome that allows channelling of the aa-tRNA from non-discriminating aspartyl-tRNA synthetase active site to the GatCAB amidotransferase site. The crystal structure of the Thermus thermophilus transamidosome determined at 3 A resolution reveals a particle formed by two GatCABs, two dimeric ND-AspRSs and four tRNAs(Asn) molecules. In the complex, only two tRNAs are bound in a functional state, whereas the two other ones act as an RNA scaffold enabling release of the asparaginyl-tRNA(Asn) without dissociation of the complex. We propose that the crystal structure represents a transient state of the transamidation reaction. The transamidosome constitutes a transfer-ribonucleoprotein particle in which tRNAs serve the function of both substrate and structural foundation for a large molecular machine.
Organizational Affiliation:
Department of Molecular Biology, CARB Centre, University of Aarhus, Arhus C, Denmark. mickael.blaise@gmail.com